Bioanalytical Research Group
Introduction to research
To create advanced and comprehensive drugs and medical technologies, it is important to understand basic underlying biological phenomena. Bioanalytical methods are indispensable for clarifying the functions and mechanisms of biomolecules such as DNA, RNA, and proteins. Our research group is developing, improving, and standardizing bioanalytical methods such as quantitative PCR, sequencing technology, and bioimaging techniques. Moreover, we are developing standard materials of biomolecules.
1: Development and standardization of bioanalytical methods
We are standardizing bioanalytical methods to facilitate the global use of biotechnology in many fields. To facilitate standardization of biotechnology, we construct a framework for cooperation with domestic industrial bodies, foreign government/research institutes, and others. The Technical Committee on Biotechnology (TC276) was established at ISO in 2013, and international standardization of cell measurement and analytical methods for biomolecules is being promoted. We develop standard materials for validation and evaluation of bioanalytical methods. These standard materials are used in the fields of food microbiology, medical engineering, and genetic testing.
2: Water-in-oil droplets technology for microbial culture
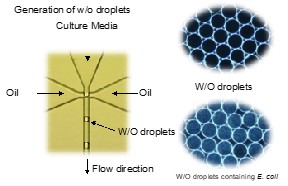 Water-in-oil droplet is a state, in which water droplets are dispersed in the oil phase. By using a dedicated device, it is possible to produce w/o droplets with a diameter of about 30 µm to 100 µm in hundreds of thousands at a time. These w/o droplets are separated by oil and can be used as independent compartments. In our research, we are trying to construct a high-throughput microbial culture method by using these w/o droplets as a culture incubator.
Water-in-oil droplet is a state, in which water droplets are dispersed in the oil phase. By using a dedicated device, it is possible to produce w/o droplets with a diameter of about 30 µm to 100 µm in hundreds of thousands at a time. These w/o droplets are separated by oil and can be used as independent compartments. In our research, we are trying to construct a high-throughput microbial culture method by using these w/o droplets as a culture incubator.
3: Bottom-up Synthetic Cell Models
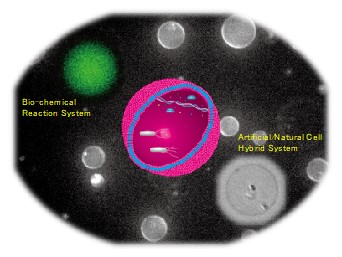 To really understand the essence of life, one approach is to realize self-organizing systems. Our aim is to construct bottom-up synthetic cell models based on vesicles, ultimately autonomous systems that can adapt their environment and evolve.
To really understand the essence of life, one approach is to realize self-organizing systems. Our aim is to construct bottom-up synthetic cell models based on vesicles, ultimately autonomous systems that can adapt their environment and evolve.
4: Development of MS-based techniques for metabolite analysis
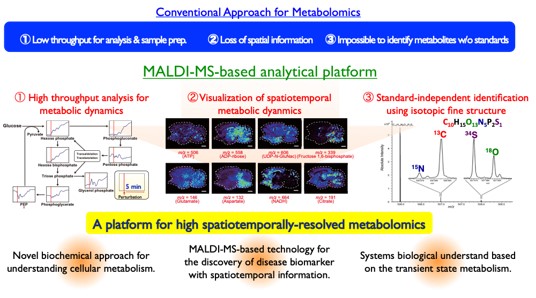 We aim to develop basic technologies and tools for quantitative and comprehensive analysis with extreme precision and resolution, which is essential for understanding the phenotype, function, diversity, and individuality of a single cell level, by sharpening our unique metabolite measurement technologies based on mass spectrometry.Especially, we have focused on the sensitivity of MALDI-MS for the analysis of low-molecular-weight metabolites and have developed a series of technologies. So far, we have succeeded in developing an high-throughput metabolic profiling technology that can be operated with high reproducibility and quantification using tens to hundreds of cells. In addition, we have succeeded in visualizing metabolic dynamics in tissue micro-regions, and developing high-precision metabolic biomarkers for cancer diagnosis using human clinical specimens.
We aim to develop basic technologies and tools for quantitative and comprehensive analysis with extreme precision and resolution, which is essential for understanding the phenotype, function, diversity, and individuality of a single cell level, by sharpening our unique metabolite measurement technologies based on mass spectrometry.Especially, we have focused on the sensitivity of MALDI-MS for the analysis of low-molecular-weight metabolites and have developed a series of technologies. So far, we have succeeded in developing an high-throughput metabolic profiling technology that can be operated with high reproducibility and quantification using tens to hundreds of cells. In addition, we have succeeded in visualizing metabolic dynamics in tissue micro-regions, and developing high-precision metabolic biomarkers for cancer diagnosis using human clinical specimens.
5: Monitoring molecular function using fluorescence correlation methods
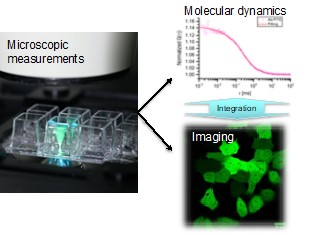 Functional biomolecules, such as nucleic acids and proteins, perform their functions by repeating their association / dissociation while changing the shape of the molecules, and moving around dynamically. However, it is not easy to directly observe the dynamics and functions of such molecules. We use fluorescence correlation methods to quantify and visualize the "function" of molecules in aqueous solutions and living cells. Also, we are developing techniques to accurately quantify the number of molecules. We believe that studying the dynamics and numbers of molecules will enable us to figure out the mechanisms of biological phenomena.We also work for assuring quantitative values in fluorescence imaging and contribute to the standardization of the bioimaging field.
Functional biomolecules, such as nucleic acids and proteins, perform their functions by repeating their association / dissociation while changing the shape of the molecules, and moving around dynamically. However, it is not easy to directly observe the dynamics and functions of such molecules. We use fluorescence correlation methods to quantify and visualize the "function" of molecules in aqueous solutions and living cells. Also, we are developing techniques to accurately quantify the number of molecules. We believe that studying the dynamics and numbers of molecules will enable us to figure out the mechanisms of biological phenomena.We also work for assuring quantitative values in fluorescence imaging and contribute to the standardization of the bioimaging field.
6: Establishment and application of measurement infrastructure for human microbiome research and development
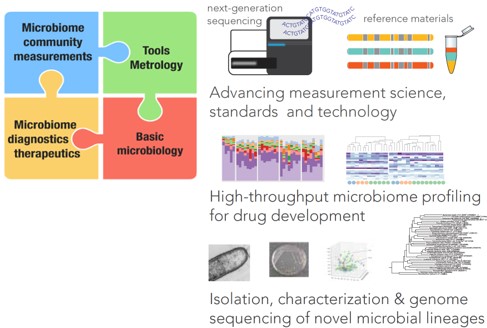 Our research is geared toward development and application of methodologies for microbiome community measurements, with focus on the human microbiome. Specifically, we are interested in establishing new metrological solutions, such as synthetic DNA/cell standards, for improving reliability of metagenomics analysis of complex microbiota, toward accelerating translation of human microbiome R&D into industrial applications to improve human health. In addition, we are exploring the relationship between gut microbiota and health and disease, with the aim of identifying key microorganisms that could serve as the basis for establishing novel therapeutics. Here, methods for high-throughput and targeted cultivation of specific strains from the human gut are also being established.
Our research is geared toward development and application of methodologies for microbiome community measurements, with focus on the human microbiome. Specifically, we are interested in establishing new metrological solutions, such as synthetic DNA/cell standards, for improving reliability of metagenomics analysis of complex microbiota, toward accelerating translation of human microbiome R&D into industrial applications to improve human health. In addition, we are exploring the relationship between gut microbiota and health and disease, with the aim of identifying key microorganisms that could serve as the basis for establishing novel therapeutics. Here, methods for high-throughput and targeted cultivation of specific strains from the human gut are also being established.
7: Understanding the mechanism of bacterial stress response
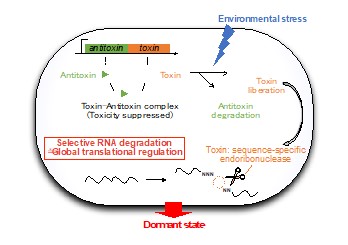 Toxin-Antitoxin system is one of the most diverse and widespread intracellular molecular mechanism of bacterial stress response. It consist of a Toxin that inhibits cell growth and an Antitoxin that, in normal condition, acts as its cognate inhibitor. When bacteria cells are exposed to environmental stresses, Antitoxins are degraded to liberate Toxins. Our research focuses on Toxins that show toxicity to the cell by cleaving intracellular RNAs in sequence-specific manners. By using massive parallel sequencing and fluorometric assay, we detect the recognition sequence of RNA-cleaving Toxins and aim to understand the bacterial translational regulations under environmental stresses.
Toxin-Antitoxin system is one of the most diverse and widespread intracellular molecular mechanism of bacterial stress response. It consist of a Toxin that inhibits cell growth and an Antitoxin that, in normal condition, acts as its cognate inhibitor. When bacteria cells are exposed to environmental stresses, Antitoxins are degraded to liberate Toxins. Our research focuses on Toxins that show toxicity to the cell by cleaving intracellular RNAs in sequence-specific manners. By using massive parallel sequencing and fluorometric assay, we detect the recognition sequence of RNA-cleaving Toxins and aim to understand the bacterial translational regulations under environmental stresses.
List of Publications
|2024|2023|2022|2021|2020|2019-2015|
Staff:
Akira Sasaki(Research Group Leader)
Naohiro Noda
Tetsushi Suyama
Akiko Yokota
Daisuke Miura
Tourlousse Dieter
Masamune Morita
Yukako Senga
Satoko Matsukura
Akinobu Nakamura