Screening, structure-function relationship analysis, and improvement of useful carbohydrate-related enzymes
For the development of industrial enzymes, we are working on the screening, analysis, and improvement of a novel enzyme. In order to explore the new enzyme, we are using not only classical microbial screening, but also metagenomic screening that can utilize genetic resources in the environment directly. We perform a detailed analysis of the enzymes to reveal the structure-function relationship, and the enzymes are then improved by rational design based on structural information and the directed evolution technique. Presently, our main targets are carbohydrate-related enzymes:
- An enzyme for saccharification of cellulosic biomass: We are studying enzymes for cellulosic biomass degradation. Cellulose, the main component of the plant cell wall, is composed of glucose. Cellulases degrade cellulose to produce glucose, which can be used in bio-ethanol production and bio-refinery. Currently, cellulase formulation derived from the filamentous fungal genus, Trichoderma, is used widely; however, it does not possess sufficient activity. Therefore, we are now developing an enzyme to compensate for the weakness of the Trichoderma cellulase formulation.
- Enzymes which degrade hemicellulose, ‘hemicellulases’ Hemicelluloses are polysaccharide components of the plant cell wall, and are structurally diverse. Hemicellulases, which degrade hemicelluloses, also display various activities in order to respond to a variety of hemicellulose structures. We are conducting research on unique enzymes that specifically recognize the structure of hemicelluloses. Enzymes that strictly recognize these sugar chain structures are useful research tools for the production of various oligosaccharide structures (which may prove useful as novel bio-based materials or as physiologically active substances), and for structural analysis of hemicellulose. Sugar chain structure-specific enzymes may prove useful as a research tool, likely in combination with restriction enzymes, for molecular biology. Several enzymes developed by us have been used as tools for the structural analysis of hemicelluloses and oligosaccharide production.
The designer of gene expression: an approach for the development of artificial transcription networks to regulate gene expression ad libitum
Transcription, the synthesis of messenger RNA from the corresponding gene locus, is one of the most fundamental biological processes in all living things. In multi-cellular organisms, thousands of genes are expressed in a single cell, but the “set of genes” expressed in one differentiated cell may be different from another. Therefore, regulation of gene expression may be synonymous with cell differentiation during development. Although the recent discovery of only four transcription factors: Oct4, Sox2, KLF3 and Myc, which can reprogram differentiated cells into induced pluripotent stem (iPS) cells, indicates that we are able to control cell differentiation ad libitum, although such regulation is difficult using current technologies. Thus it is necessary to study deeply and understand the complex interaction between cis- and trans-acting factors. For example, the structure of basal, or core promoters, as engines of the transcription machinery of eukaryotes (which is composed of the common cis-elements and their corresponding trans-acting factors) has been well-documented because it is evolutionarily conserved and located close to the transcription start site of the corresponding downstream gene. In most cases, however, the principle of cell-type specific gene expression is still incomprehensible because such cell-type specific enhancers (or silencers) were hidden in the illimitable woods of genome sequences. The recent revolution in sequencing technologies spawned a huge quantity of whole genome data in a short period of time at a reasonable cost. With comparative genome analyses utilizing new sequencing technologies and bioinformatics, we hope to elucidate the principle of tissue- or cell type- specific gene expression in multi-cellular organisms. Our goal is to establish a novel discipline, “promoter engineering,” in order to provide a framework for designing artificial transcription systems. We have proceeded with the following projects:
- Development of computational analysis by comparing regulatory regions between microbes and man using the massively parallel next-generation sequencer.
- Characterization and modification of small, addorsed bidirectional (SAB) and cell-type specific (e.g. neuron-specific) promoters by comparing regulatory regions between related species which have counterfeit genomes of the Japanese rice fish Medaka (Oryzias latipes).
- Implementation and verification of artificial transcription networks introduced into cells or living organisms designed to optimize gene expression ad libitum by combining modified genes, cis-acting regulatory regions, trans-acting factors, and non-coding RNAs.
Development of gene expression systems for the production of useful materials in yeast
Expression systems in eukaryotic microorganisms are useful for the production of various proteins (including human proteins), with respect to a greater probability of proper folding compared with Escherichia coli expression systems. The budding yeast Saccharomyces cerevisiae is a good host for eukaryotic expression systems, since various methods of genetic engineering are available, and well-organized genome information is available. Only a few genetic elements (e.g. promoters and signal peptides) are utilized in existing yeast expression systems; however, a large number of genetic elements in the S. cerevisiae genome remain unused. Therefore, we want to develop efficient expression systems for the production of useful materials by yeasts using the various genetic elements contained in the yeast genome.
- Development of efficient protein expression systems in yeast using its genomic information: In order to provide a useful expression system for human and mammalian proteins, we are constructing a unique and powerful low temperature S. cerevisiae expression system at. This system is optimally designed based on the comprehensive analysis of gene expression in yeast, as well as bioinformatics using yeast genome information. When enhanced green fluorescent protein (GFP) was expressed in this system at a low temperature, its volume reached approximately 50% of the soluble protein in yeast cells, surpassing the existing normal temperature yeast expression systems. Furthermore, we confirm that our expression system can produce many human proteins in a soluble form, which is difficult using existing expression systems. We are now improving our expression system to produce various other proteins.
- Development of expression systems for efficient xylose metabolism in bioethanol-producing yeast: S. cerevisiae has naturally high ethanol production ability, and is widely used for ethanol production from glucose. However, wild-type S. cerevisiae cannot ferment xylose, which is the dominant pentose sugar in plant biomass hydrolysates. One strategy to engineer yeasts capable of efficiently producing ethanol from xylose is by introducing the genes involved in xylose metabolism. However, optimization of the expression levels of xylose metabolic enzyme genes for efficient xylose metabolism in yeast has not been investigated in detail. Therefore, we want to improve the xylose to ethanol conversion efficiency in yeast by controlling expression levels of xylose metabolic enzyme genes using various promoters in the yeast genome. We are also attempting to develop expression systems for conferring thermo- and low pH-tolerance to a bioethanol-producing yeast by comparative genome analyses of thermo- and low pH-tolerant yeast strains using the next-generation sequencer.
The role for glycosphingolipids in adaptive immune response and its application to industrial technology
Glycosphingolipids (GSLs) are cell membrane components composed of oligosaccharides and ceramides. Oligosaccharides and ceramides in GSLs are structurally diverse, and recent studies have revealed that GSLs containing very-long-chain fatty acids (VLCFAs) in the ceramide portion are involved in immune responses in mammalian tissues. Our study focuses on the role for GSLs in adaptive immune response and the effect of diets on tissue GSL metabolism.
(1) Induction of adaptive immune responses by glycosphingolipids containing very-long-chain fatty acids.
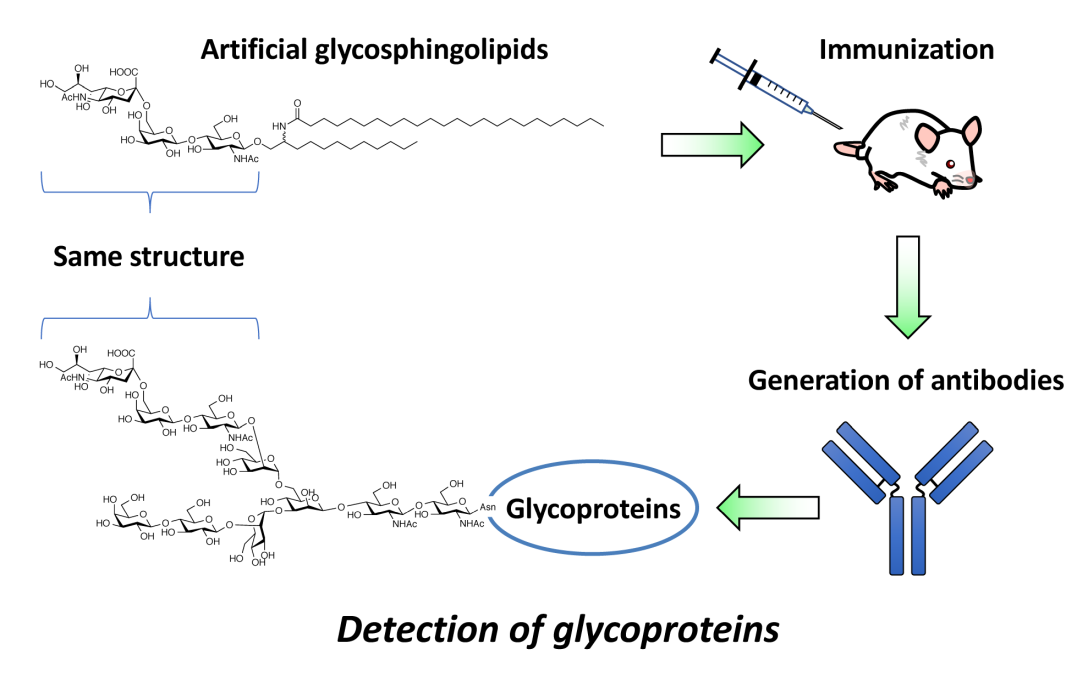
GSLs containing VLCFAs regulate several immune responses, such as cytokine production, immune signaling, and antibody induction. We found that immunization with GSLs containing VLCFAs can efficiently induce the production of anti-glycan antibodies by B cells. Based on this finding, we have developed artificial GSLs which can specifically induce adaptive immune responses and promote antibody production by B cells. The technology can be utilized to develop anti-glycoprotein antibodies and cancer vaccines targeting tumor-associated carbohydrate antigens [Ref.1].
(2) Diet and Ganglioside Expression.
Gangliosides are series of GSLs containing sialic acids in the oligosaccharide portion in mammalian cells. Gangliosides are a component of cellular membranes and play roles in modulating membrane function and the activity of membrane proteins. Abnormal expression and metabolism of gangliosides lead to the onset of several conditions in humans, such as neurologic diseases, diabetes, and cancer. A number of studies have been carried out to date to investigate the role of gangliosides in these diseases, and the effect of diet on tissue expression of gangliosides has recently become a topic of interest in this field. As gangliosides are degraded in the intestinal tract, ingested food-derived gangliosides are not directly absorbed into tissues in vivo, but the degradation products can be absorbed and affect ganglioside expression in the tissues. Recent studies have also shown that the expression of gangliosides in tissue cells can be indirectly induced by controlling the expression of ganglioside metabolism-related genes via the diet. Our study focuses on the dietary control to regulate the expression levels of gangliosides in tissues, which is expected to play a role in preventing and treating ganglioside-related diseases[Ref.2].
- Induction of specific adaptive immune responses by immunization with newly designed artificial glycosphingolipids. Sci. Rep. 2019, 9, 18803.
- Dietary Control of Ganglioside Expression in Mammalian Tissues. Int. J. Mol. Sci. 2020, 21(1), 177.
References
Elucidation of molecular mechanisms of sophisticated social systems in insects
Social insects have evolved sophisticated social organization in their colony. In our study, we are focusing on ants and aphids to clarify molecular mechanisms of their highly specialized biological functions to maintain and control the social systems. These studies using such unique animals are expected to discover novel biological phenomena and functions, bioactive substances, and lead compounds for medical drugs etc.
(1) Molecular mechanisms of caste differentiation, social behavior and insect-plant interactions in social aphids.
In some aphids, altruistic individuals known as “soldiers” are found in their colonies like bees, ants and termites. Among about 5,000 aphid species recorded in the world, more than 80 species are known to be social. Their primary social role is colony defense, but soldiers in some species also perform nest labors, such as gall cleaning and repairing. Soldiers and reproductive individuals are genetically identical clones produced by parthenogenesis, but their morphology, reproductivity and behavior are totally different because of their caste-specific gene expression patterns. We are focusing on soldier aphids to elucidate molecular mechanisms of caste differentiation and social behaviors in aphids. We are also interested in aphid-plant interactions, such as gall formation by social aphids. Also see our publications.
(2) Regulatory mechanisms of social behavior in ants
Social animals live in groups with other members. With social ants, our goal is to understand the relationship between our health, longevity, and social environment with utilizing the behavior tracking system, omics analysis, and molecular biology.
Social insects live in the social organization called colony, with complex social hierarchy composed from reproductive queens, males and non-reproductive female workers. In matured colony, workers have another sociality called division of labor in worker castes. Workers perform nursing when they are young, and shift their job to foraging when they get old. They also can flexibly change their job depending on the demand in their colony. We focus on their sophisticated social organization and study the molecular mechanisms to regulate their behavior, physiology and longevity in social environment. See here for the details.