Current main research topics
To solve various problems like environmental protection, energy and food shortage and so on and to provide healthier and happier life, we are engaged in the development of new technologies to enhance plant ability by utilizing our own original gene regulation techniques.
1. Development of new bioresource plant to contribute bioeconomy
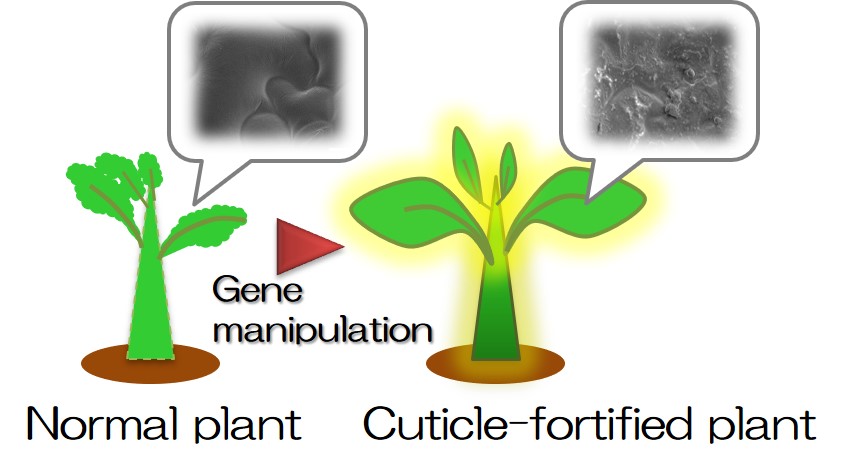
Plant is a renewable bioresource and inevitable for bioeconomy. We are developing new bioresource plant more suitable for its exploitation by employing our advanced technologies for the analysis of cell wall and woody substance, which is the world's largest terrestrial biomass, and surface cuticle, which protects plant (Figs. 1, 2).
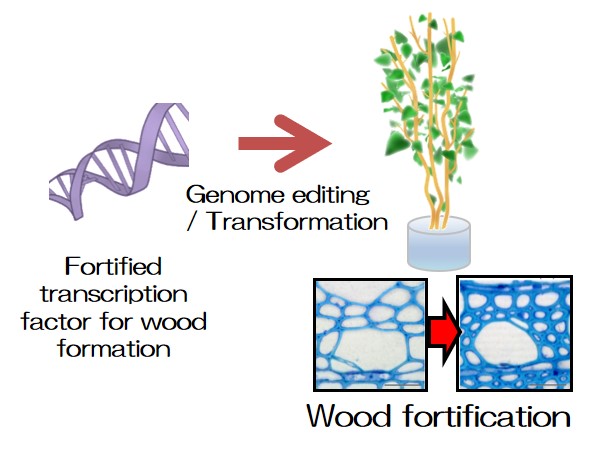
- Fig. 1: Fortifying wood formation by gene regulation
- Fig. 2: Fortifying cuticle by gene regulation
2. Development of new resilient plant against climate change
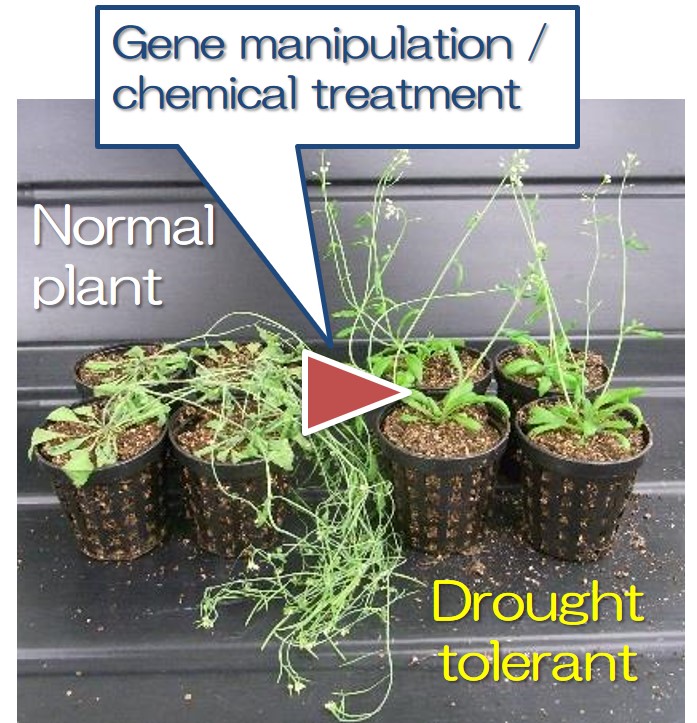
Climate change due to global warming has been impacting on crop productivity in recent years. We are developing new resilient plant with more tolerance to drought, heat and pathogen attack by our original manipulation technique of gene regulation (Fig. 3).
- Fig. 3: Increasing drought tolerance by genetic manipulation or chemical treatment
3. Development of plants making us healthier and happier
Plant is not only important for resources and food but also important for increasing our health and healing our mind. We are developing new useful plants with increased production of a particular beneficial ingredient or new ornamental plants with excellent characteristics through our techniques finding promising genes (Fig. 4).
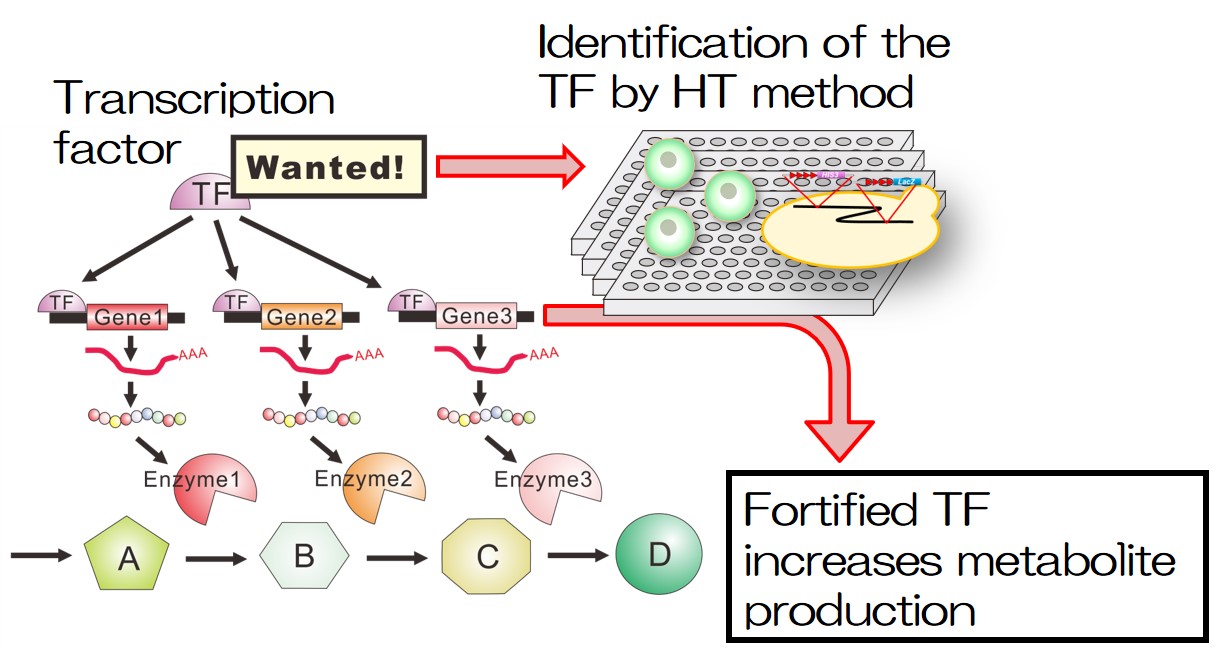
- Fig. 4: Improving metabolite production by finding and regulating transcription factor
4. Development of new genome-editing technology for the rapid application of our outcome to the society
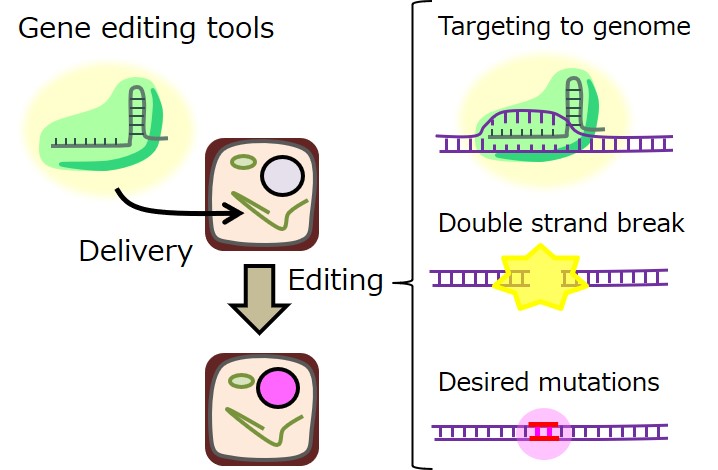
To widely prevail our newly-developed plants, more socially-acceptable form is required to provide. We are going to develop new genome-editing technologies enabling us to do more flexible but human and eco-friendly genome-edit to realize our dream (Fig. 5).。
- Fig. 5: Targets of technical development to improve genome editing technology