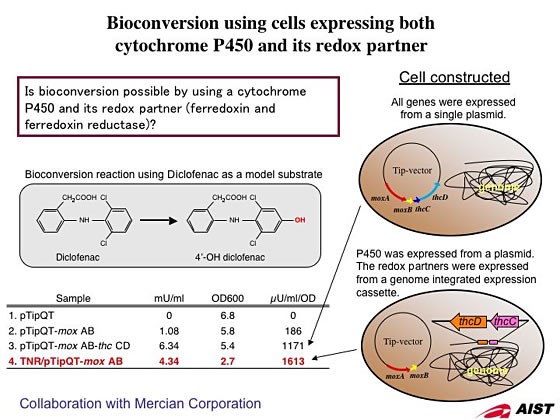
1. Development of bioconversion techniques using Rhodococcus as a host.
We have been developing two different protein expression systems using Rhodococcus as a host; one is expression vector that have an autonomous replication origin and another is genome-integrating expression system. The expression vectors have a variety of combinations including two different replication origins, selection markers, and two different promoters that can be selected from constitutive and inducible type. By using these vectors that have been developed in our lab, we confirmed that proteins could be expressed over a wide range of temperature from 4 to 35°C. The genome-integration system has been developed by using transposon vector modified from IS1514. This expression system does not need the addition of antibiotics into the culture media, which are indispensable in case of expression vectors, and it is suitable for an industrial scale culture.
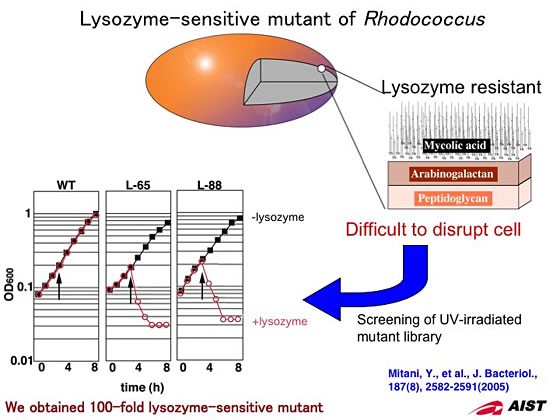
2. Screening for lysozyme-sensitive mutant.
The cell wall of Rhodococcus composes peptidoglycan, arabinogalactan, and mycolic acid. This cell wall structure makes Rhodococcus resistant to lysozyme, which is widely used to lyse cells when extracting cellular components. We screened for lysozyme-sensitive mutants after UV irradiation to wild type cells and successfully cloned two lysozyme-sensitive mutants from over 8500 colonies. These mutants showed 100-fold sensitivity to lysozyme and were lysed immediately after the addition of low concentration lysozyme. By using these mutants, we have dramatically improved the protein extraction process.
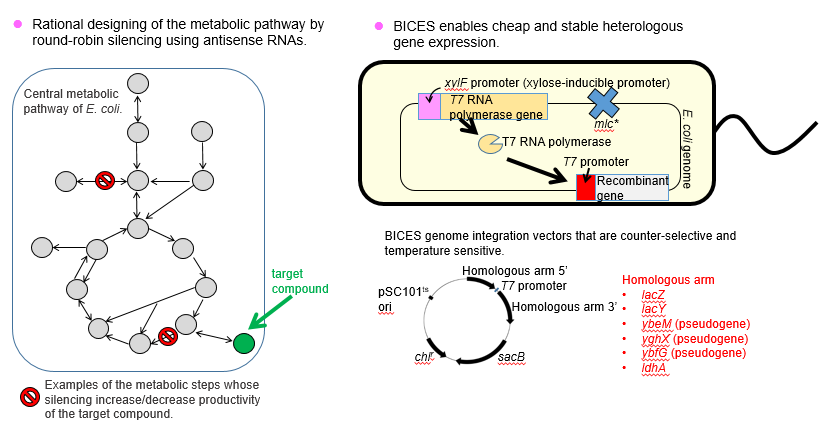
3. Design the metabolic pathway of E. coli "reasonably" and modify it "inexpensively"
Production of industrially useful compounds using microbes such as Escherichia coli is considered to contribute to breaking away from the petroleum-dependent society and constructing a sustainable society. However, since natural E. coli cells do not produce useful compounds, the metabolic pathway must be modified by gene recombineering techniques.
So far, researchers have investigated the optimal metabolic pathway to produce target compounds by trial and error which is based on the experience and knowledge of the researcher and is painful and time-consuming. Therefore, to design the optimal metabolic pathway rationally and easily (= without repeating wasteful trial and error), we developed a novel design approach in which antisense RNA is used to silence gene expression. Specifically, we created 71 antisense RNA expression vectors that silence all metabolic steps of the central metabolic pathway. By conducting this round-robin silencing, we could know the metabolic steps that had a critical effect on productivity and design the optimal metabolic pathway from that information.
After knowing the optimal metabolic pathway, the metabolic genes must be modified as designed. To this end, introduction and expression of heterologous genes are frequently necessary, and we developed a new method to do so; in the method, many heterologous genes are inserted into the E. coli chromosome and expression of these genes is induced with xylose contained in biomass as raw material. This method is named BICES (Biomass Inducible Chromosome-based Expression System). BICES enables to express heterologous genes stably over a long period of time without using expensive expression inducing agents (such as IPTG) or antibiotics for plasmid maintenance.
Using unique methods as above as well as other conventional methods, we have succeeded in producing acetoin, 2, 3 butanediol, pyruvic acid, isobutanol and so on in E. coli.
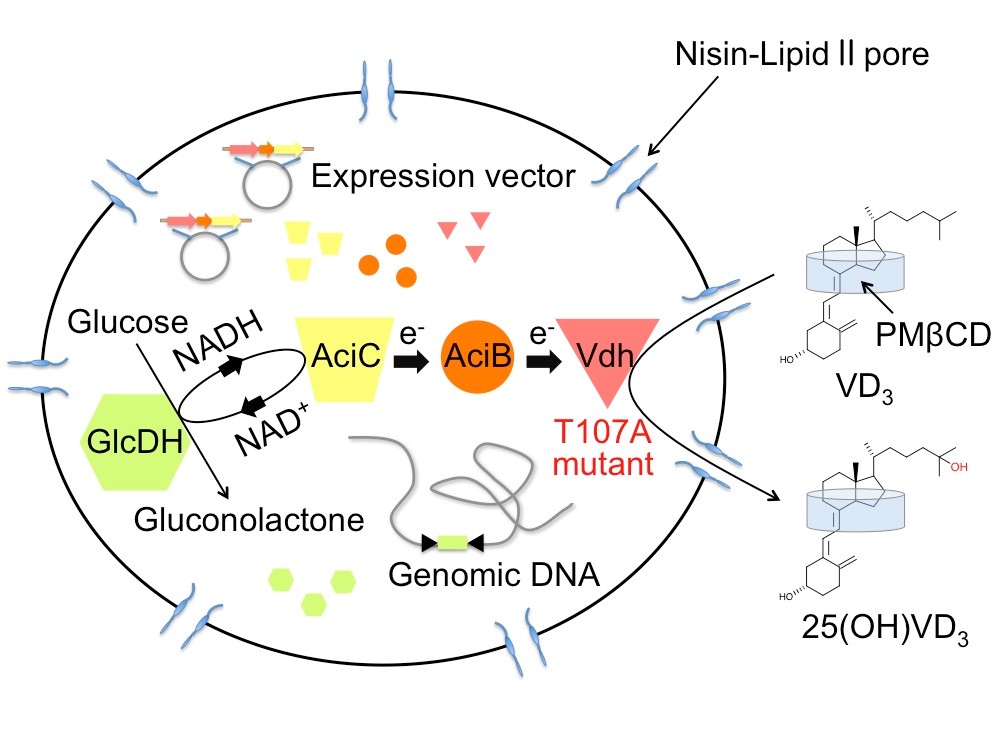
4. Bioproduction of hydroxyvitamin D3 by nisin-treated Rhodococcus erythropolis cells
Pseudonocardia autotrophica is able to convert from vitamin D3 (VD3) to hydroxylated VD3, the reaction of which is catalyzed by cytochrome P450 vitamin D hydroxylase (Vdh). Hydroxylated VD3 is useful as a pharmaceutical for the treatment of conditions associated with VD3 deficiency and VD3 metabolic disorder. We have determined three-dimensional structure of Vdh by X-ray crystallography to understand the substrate binding and catalytic mechanisms. We also created highly active Vdh mutants by structure-based and directed evolution studies. In addition, we have developed highly efficient hydroxylated VD3 production system using nisin-treated Rhodococcus erythropolis cells expressing the highly active Vdh mutant.
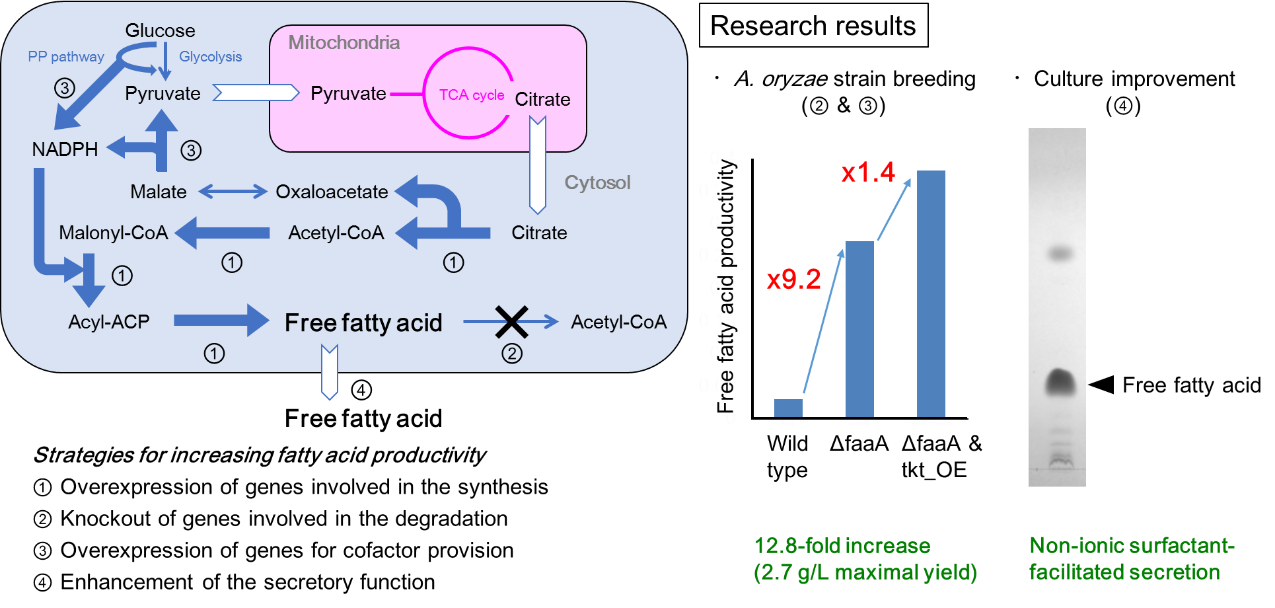
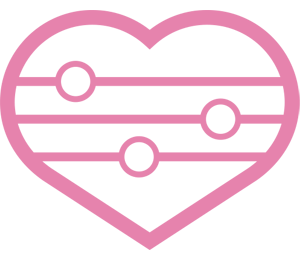
5.Increase of free fatty acid productivity by metabolic engineering of Aspergillus oryzae
Free fatty acids (FFAs) are useful as source materials of pharmaceuticals, biofuel, etc. We could increase FAA productivity at 12.8-fold by engineering the fatty acid metabolism in A. oryzae. It corresponds to production of 2.7 gram FFA per liter for 5 days. Also, by supplementing non-ionic surfactant to the culture, over 80% of FFAs were found to be released extracellularly without inhibiting the cell growth. By the finding, FFAs could be released easily without much labor. Hereafter, by introducing further metabolic engineering to A. oryzae, we are aiming at producing FFAs whose structures are modified to be similar to pharmaceuticals etc. Throughout the research, we would like to apply our FFA production technology by A. oryzae to industrial use.