Our research focuses on producing useful compounds using microorganisms. As microorganisms, Escherichia coli, a kind of an actinomycete Rhodococcus erythropolis, and koji mold are used. The target compounds to be produced are pharmaceutical materials, petroleum alternative fuels, and fine chemicals, etc. In addition to manipulating microbial cells, our study also involves analyzing and utilizing information on protein structures and genome information. Besides, we are developing functional evaluation of foods and bioassay systems using microorganisms and cultured cells.
1. Development of protein expression systems.
We are developing expression vectors suitable for producing valuable proteins in a large scale by using Rhodococcus as a biofactory or for accumulating desirable proteins to create a multi-functional cell. In particular, we aim at efficient protein production systems by searching for inducible or constitutive strong promoters that control the expression of target genes.
2. Development of techniques to manipulate cell wall structure.
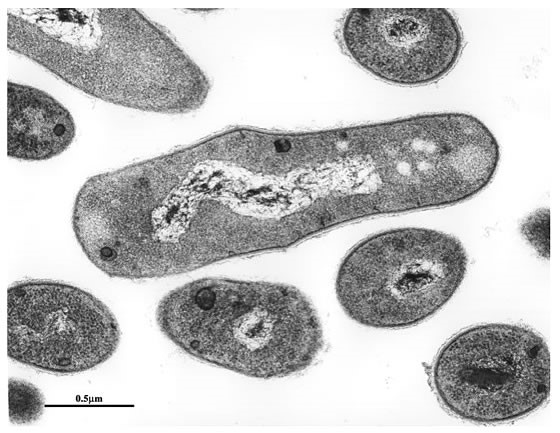
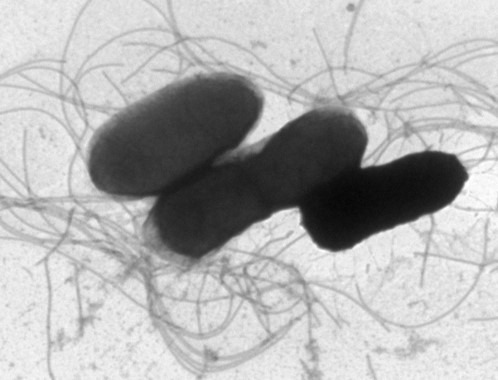
Rhodococcus cells have a characteristic cell wall structure that contains peptidoglycan, arabinogalactan, and mycolic acid. The cell wall structure confers Rhodococcus cells a hard shell that is resistant to physical and chemical attacks from the outside and it functions as a permeability barrier. In order to extract proteins expressed in the cell, we have to break the robust cell wall. For this purpose, we obtained mutant strains that are easily disrupted by chemical treatment in a mild condition. Also, we are trying to obtain and analyze mutant cells with higher cell permeability that will make it possible to convert substrate supplied in the medium more efficiently.
3. Producing valuable compounds using Escherichia coli.
E. coli is one of the most frequently used microorganisms to produce various useful compounds microbially, because it is easy to handle genetically. We have developed unique technologies such as a rational metabolic engineering method using antisense RNA and a heterologous gene expression method in which expensive gene expression inducer and antibiotics are not necessary (BICES; Biomass Inducible Chromosome-based Expression System) which does not require antibiotics. We have succeeded in engineering the metabolic pathway of E. coli using these methods and producing various useful compounds (#Link: To the page of the former member Nobutaka Nakashima senior researcher) . For example, we succeeded in producing acetoin, 2,3- butanediol, pyruvic acid, isobutanol and etc.
4. Producing valuable compounds using Aspergillus oryzae.
Aspergillus oryzae secretes large amount of hydrolytic enzymes, while does not produce any toxins. Therefore, it has been used for manufacturing Japanese traditional foods such as alcohol (sake), soy sauce (shoyu), and soy paste (miso). A. oryzae is the filamentous fungus that has organelles in the cell, and thus it is considered appropriate for heterologous expression of genes derived from plants and animals, etc. Using such attractive characteristics of this fungus, we have been challenging the production of valuable compounds such as lipids and secondary metabolites useful as source materials of pharmaceutical agents etc. via genetic engineering.
5. Exploration for bacteria producing valuable compounds.
In actinomycetes, many Streptomyces species are known to produce antibiotics. We are exploring bacteria, other than Streptomyces, that are producing compounds toxic to other bacteria and are trying to determine the nature of these compounds. Such compounds will be useful as antibiotics. Also, by analyzing the synthetic pathways of these compounds, it will be possible to produce such compounds in a large scale.
6. Development of techniques to improve enzymatic function.
It will be possible to make cells that have improved ability by expressing small amount of high performance enzyme, which may have 10-100 fold activity than the original one, instead of a high expression of normal enzyme. It is also possible to improve reaction efficiency by changing substrate specificity and/or protein stability. For this purpose, we are developing basic techniques to improve protein function by analyzing atomic-resolution crystal structure of useful proteins.
7. Development of techniques to control protein stability in cells.
In vivo, proteolysis is subject to spatial and temporal control in order to prevent damage to the cell. By understanding the protein degradation mechanism, it will be possible to control the half-lives of proteins and to make proteins accumulate in cells efficiently. In this background, we are studying on the protein degradation system by using a proteome analysis approach. We are also studying on the protein degradation systems of higher animals and exploring proteins whose expression levels are influenced by disease or aging. These proteins can be used as molecular markers for diagnosis.
8. Development of functional lipid production and their application.
We are working on improving polyunsaturated fatty acid (PUFA) productivity of microorganisms and on developing technologies to efficiently acquire PUFAs. PUFAs such as docosahexaenoic acid (DHA, 22:6) and eicosapentaenoic acid (EPA, 20:5) are among the essential nutrients, which can hardly be synthesized in vivo. These PUFAs are known to reduce the risk of coronary heart disease and to alleviate inflammatory diseases. Then, PUFAs are expected to be used as health foods and pharmaceutical raw materials. We also aim to advance the biotechnological methods of PUFA-containing lipid production using biosynthesis genes for PUFA obtained from microorganisms and algae.
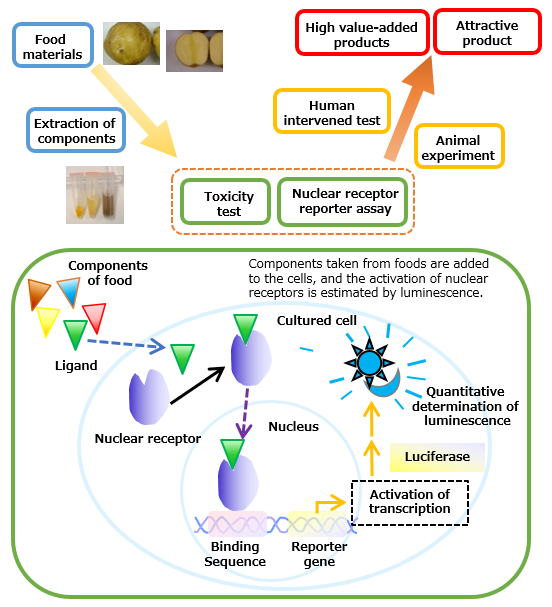
9. Functional analysis of food materials and their application by evaluation of nuclear receptor activation.
We have constructed a "nuclear receptor reporter assay" by introducing a luciferase gene linked downstream of the DNA sequence to which the activated nuclear receptor binds in cultured cells. When the food component is an agonist, or a substance showing various physiological actions due to binding with a receptor, of nuclear receptors, activation of transcription occurs and luciferase is synthesized. Therefore, it is possible to evaluate the amount of nuclear receptor activation by quantifying the amount of luminescence by luciferase.
In addition, we are developing more convenient nuclear receptor reporter assay system that is easier to use, in order to increase the throughput of "nuclear receptor reporter assay".
10. Bioinformatics analysis of environmental microbial community.
In our daily lives, such as in soil and water, on the body surface of organisms, and in the gastrointestinal tract, there are various biological communities composed of many microbial species. Many microorganisms, including hard-to-cultivate microorganisms, coexist in these communities, and most of their genetic information and ecological relationships are still poorly understood. We think that findings gained from these microbial community analyses contribute to developing novel bioproduction technologies. Therefore, we are developing bioinformatics methods to explore the unknown function of genetic resources and ecological relationships in environmental microbial communities.