1. Culturing the uncultured fastidious microorganisms in the environment
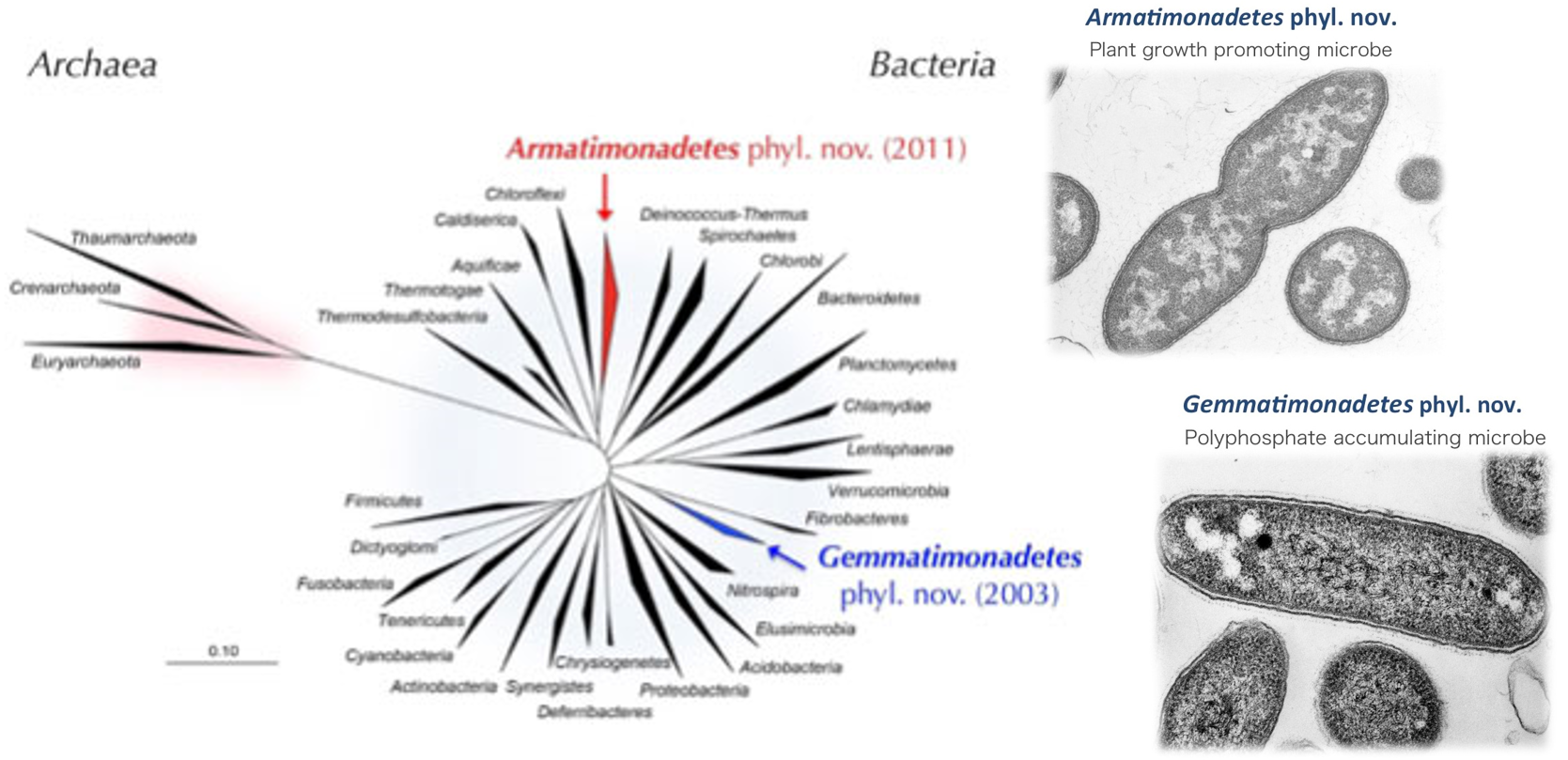
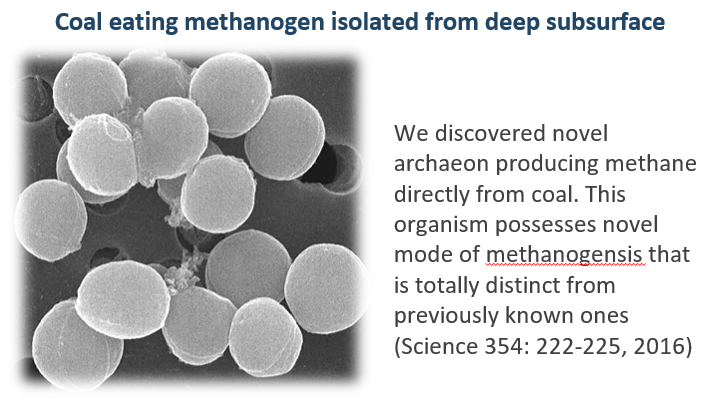
To search for uncultured microorganisms, we have developed new strategies to isolate microorganisms that resist conventional cultivation. Using those methods, we have succeeded in isolating a number of yet-to-be cultured microorganisms from various natural and artificial environments including soils, freshwater sediments, hot springs, composts, activated sludge, deep subsurface waters and sediments. Indeed, novel taxonomic names of 65 species, 37 genera, 11 families, 8 orders, 5 classes, and 2 phyla have been proposed since 2000. In particular, we isolated a new phylum bacterium, Gemmatimonas aurantiaca, and proposed a new phylum, Gemmatimonadetes in 2003. We further succeeded in isolating another new phylum bacterium, Armatimonas rosea, belonging to the formally called candidate phylum OP10 that was one of the most widely recognized uncultured phyla. A novel bacterial phylum, Armatimonadetes, was approved by International Committee on Systematics of Prokaryotes (ICSP) in 2011. Recently, we isolated novel microorganisms with unique functions: a type II diabetes inducing intestinal Lachnospiraceae bacterium, Fusimonas intestini gen. nov., sp. nov., a coal-eating deep subsurface methanogen with the novel mode of methanogenesis (Science 2016). In 2020, JAMSTEC and our group in AIST is successful of isolating a novel archaeon (belongs to so-called “Asgard” archaea superphylum) at the prokaryote–eukaryote interface and proposes a new evolutionary model for eukaryogenesis (Nature 2020).
2. Exploration and elucidation of unidentified functions in novel biological and genetic resources and their application for bio-industries
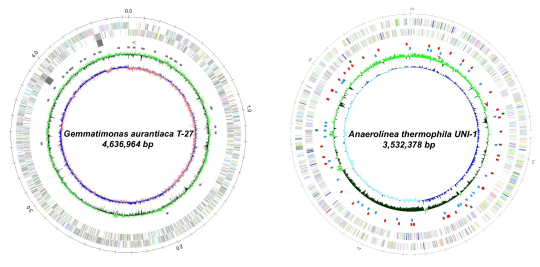
Using novel microbial and genetic resources obtained in our researches, we have attempted to create new bio-products in extensively collaboration with a wide variety of universities and private companies with strong expertise in bioindustry and environmental technology. In addition, whole genome analysis of newly isolated microorganisms with high novelty or unique functions has been conducted. To date, the genome analysis for Gemmatimonas aurantiaca (a new phylum bacterium), Armatimonas rosea (a new phylum bacterium) Caldlinea aerophila (a new class bacterium), Anaerolinea thermophila (a new class bacterium), and Microlunatus phosphovorous (a polyphosphate accumulating bacterium) has been completed.
We also undertake research on cell-cell communication (Quorum Sensing) and disruption of quorum sensing (Quorum Quenching). We have succeeded in isolating novel quorum-quenching bacteria/enzymes capable of interfering quorum sensing system composed of competitive bacteria. Furthermore, we have discovered that a novel quorum-quenching enzyme isolated from multidrug resistant bacterium was further able to degrade antibiotics and confer multidrug-resistance as well as quorum quenching on the host organism, indicating that the new bifunctional enzyme mediates two different biological functions. In this way, we are trying to reveal new microbial functions through the characterization of hitherto-unknown enzymes. In addition, we also aim to explore genetic/enzymatic resources leading to industrial uses.
3. Omics-driven discovery of novel genetic resources
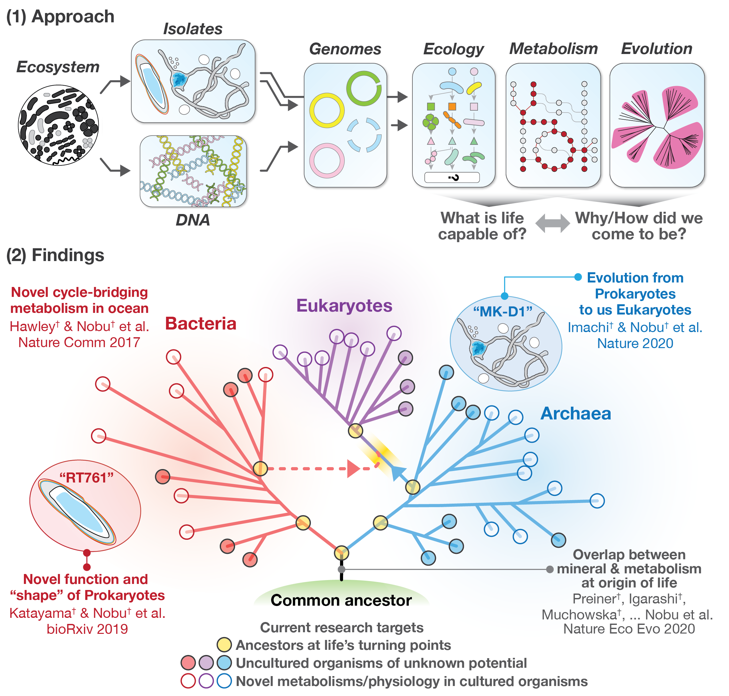
Without understanding of the hidden biochemical prowess of life, we cannot expect to develop innovative technologies that are essential for environmental stewardship/sustainability and our health. As emergence of environmental, social, and medical issues accelerate at an alarming rate, we must efficiently acquire new and applicable knowledge to push the frontiers of current biology and biotechnology forward. One invaluable and poorly explored biological resource is the plethora of uncharacterized genes that exist in nature, from which we can decipher the biochemical potential and history of life on Earth. The group combines precise environmental genome analysis (metagenomics, metatranscriptomics, etc.) with innovative cultivation techniques to unravel novel metabolic functions and life’s history from genes and genomes.
More than 90% of the microorganisms on Earth remain uncharacterized and their functions are unknown. In addition, more than 400 million genes have been discovered through research on various microorganisms and genes, but less than 1/4,500 of them have been characterized with function. We perform advanced genome analyses synthesizing environmental genomics, biochemistry, thermodynamics, evolution, and machine learning to predict and reconstruct novel metabolic functions and life’s evolutionary history. Through synergistic collaboration with culture specialists, we have been able to investigate the untapped abilities of cultured microorganisms and also accomplish genome-inspired cultivation of uncultured microorganisms.
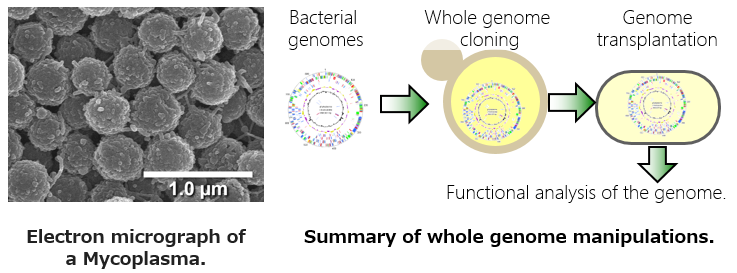
4. Development of whole genome manipulation techniques
There is huge variety of microorganisms in nature. It has been known that most bacteria in nature are unculturable. The proportion of culturable bacteria are thought to be less than 1% of natural bacterial variety. Moreover, some symbiotic and pathogenic bacteria could not be cultured anymore because of extreme genome reductions, loss of a lot of genes by adapting nutrient-rich environments. Thus, unculturable bacteria would be unexplored areas, and many researches are recently focusing on unculturable bacteria.
Recently novel methods for genome manipulation have been developed. Our group focuses on development of new techniques of genome manipulation and tries to apply them to the unculturable bacterial researches. In particular, we have attempted to clone a whole genome of an uncultured microorganism into an easily cultured microbial host, and to reveal the unknown biological functions in the uncultured majority.
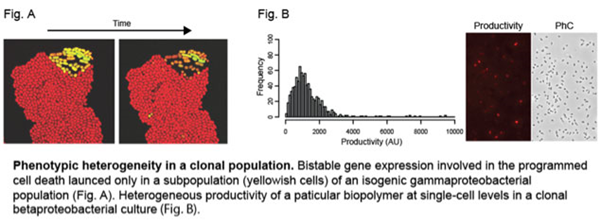
5. Functional and evolutionary aspects of phenotypic heterogeneity in a clonal bacterial population
In a classical way, clonal bacterial populations have been thought of as millions of uniform cells, exhibiting little or no phenotypic diversity. However, recent studies using single-cell based approaches revealed such an idea is just an oversimplification. Rather, a pool of isogenic bacterial cells can contain phenotypically distinct subpopulations, which may arise either from stochastic variation in several physiological processes or deterministic reactions such as feedback regulations.
Our goal is to unveil molecular mechanisms underlying phenotypic diversity and reveal physiological functions and evolutionary roles of such heterogeneity in a clonal population. To address the questions, we use molecular genetics and single-cell live imaging techniques combined with high-throughput technologies. Furthermore, we would propose a novel bioengineering scheme based on such cellular potentials at single-cell level.
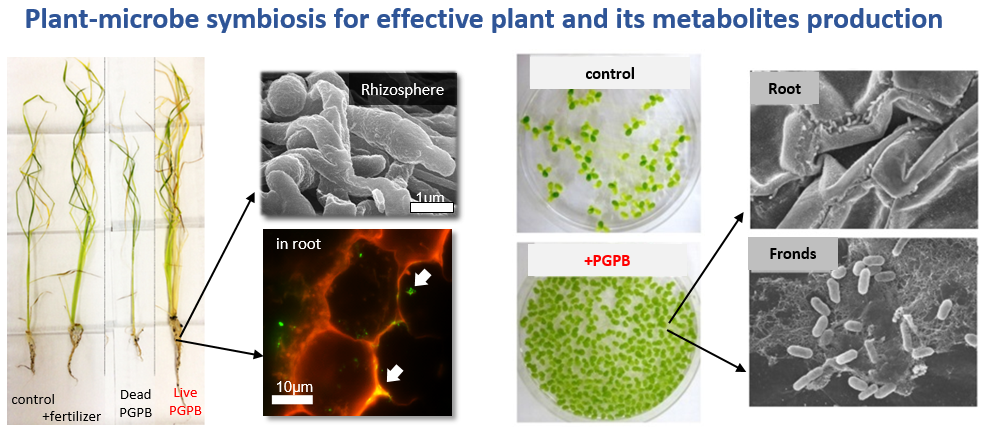
6. Ecophysiology of environmental microorganisms contributing to energy production and environmental purification
By the combined use of the cultivation techniques and the advanced molecular ecological methods via next generation sequencing technology, we aim to elucidate ecophysiological functions of environmental microorganisms associated with energy production and environmental purification. In particular, we have focused on symbiotic interactions between aquatic plants and their root microbes since the plant-microbe interactions may contribute to development of novel bioprocess for effective environmental control and bio-product innovation (supported by JST-ALCA: Advanced Low Carbon Technology R&D). So far, we have found novel microorganisms that can vigorously promote growth of the aquatic plants (PGPR: plant growth promoting rhizobacteria) and attempted to elucidate the underlying mechanisms of the novel biological interactions. We also focus on novel microbiological processes behind the biological sink of atmospheric trace gases. In particular, plant-associated bacteria demonstrating the ability to consume atmospheric H2, the tropospheric burden of this indirect greenhouse gas is considered to increase under a future H2-based economy, is investigated to determine their phylogenetic identity and eco-physiological properties. We aim to provide useful findings for technology development of environmental engineering consulting atmospheric composition change.
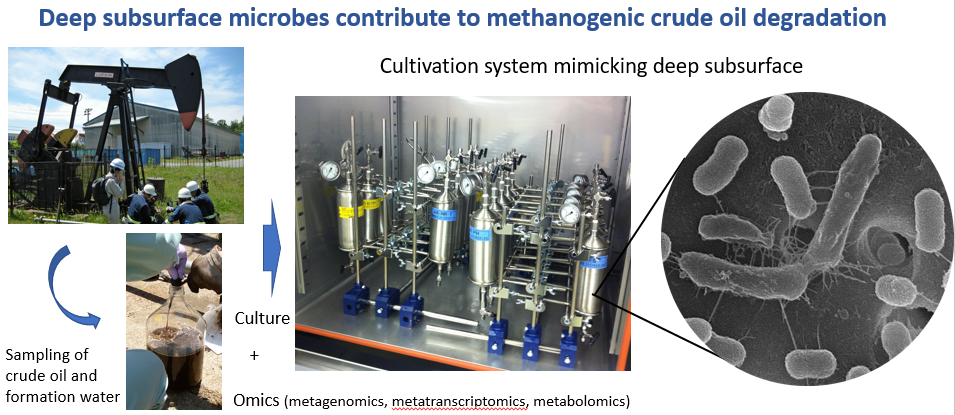
Furthermore, we have investigated anaerobic microbial communities and functions in terrestrial subsurface environments because of their great impact on global carbon cycling and energy production such as methane production. In particular, we have conducted the collaboration research with Geomicrobiology Research Group (http://unit.aist.go.jp/georesenv/geomicrob/member_en.html) in Institute for Geo-Resources and Environment (GREEN) and several private companies to better understand microbial communities and their ecophysiology associated with methanogenic crude oil degradation in the depleted oil reservoirs. Through the research, we aim to provide new insights into geomicrobiology in the oil fields and contribute to development of novel enhanced oil recovery (EOR) technologies.
7. Physiology and eco-evolutionary dynamics of host-microbe interactions
Interaction between animals and microorganisms has a great influence on the host physiology, such as health and disease, and thus is attracting attention both socially and industrially. It is also known that animals and microorganisms that have a symbiotic relationship co-evolve by interacting with each other to enhance their fitness.
Using mammals and insects as research models, we study various physiological phenomena, signal transduction, and metabolic networks that occur due to the interaction and symbiotic relationship between hosts and microorganisms. In addition, we study the co-evolution of hosts and microorganisms and their eco-evolutionary dynamics in nature. Using social insects, we aim to understand how gut microbial communities influence the host society and what principles underlie microbial community formation.