生物データサイエンスグループ
研究概要
ウエット研究者とドライ研究者が共存していることが本研究グループの大きな特徴です。インシリコによる創薬や診断を支援する独自技術の開発を行うだけでなく、ウエット研究とドライ研究のシナジー(独自の計測技術や産生法により得られたオミクス情報や治療モダリティを理論計算や情報解析技術により高度化すること)により、創薬・診断・治療をブーストする技術の開発を行い、それらの技術の社会実装を目指しています。また、ウエット研究とドライ研究の両方を行える人材の創出も取り組んでいます。
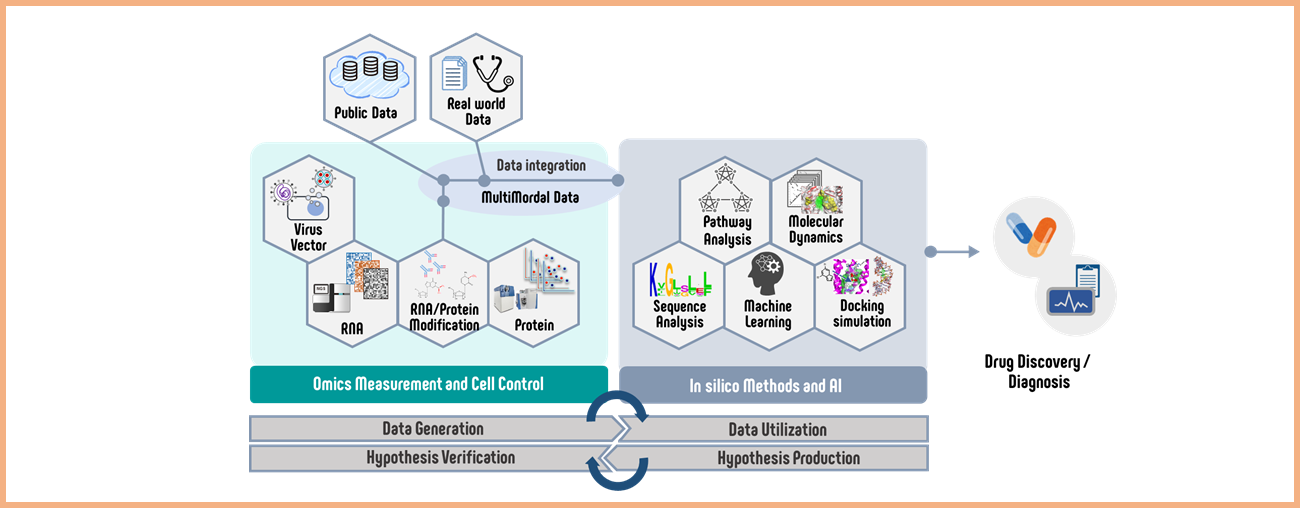
研究課題
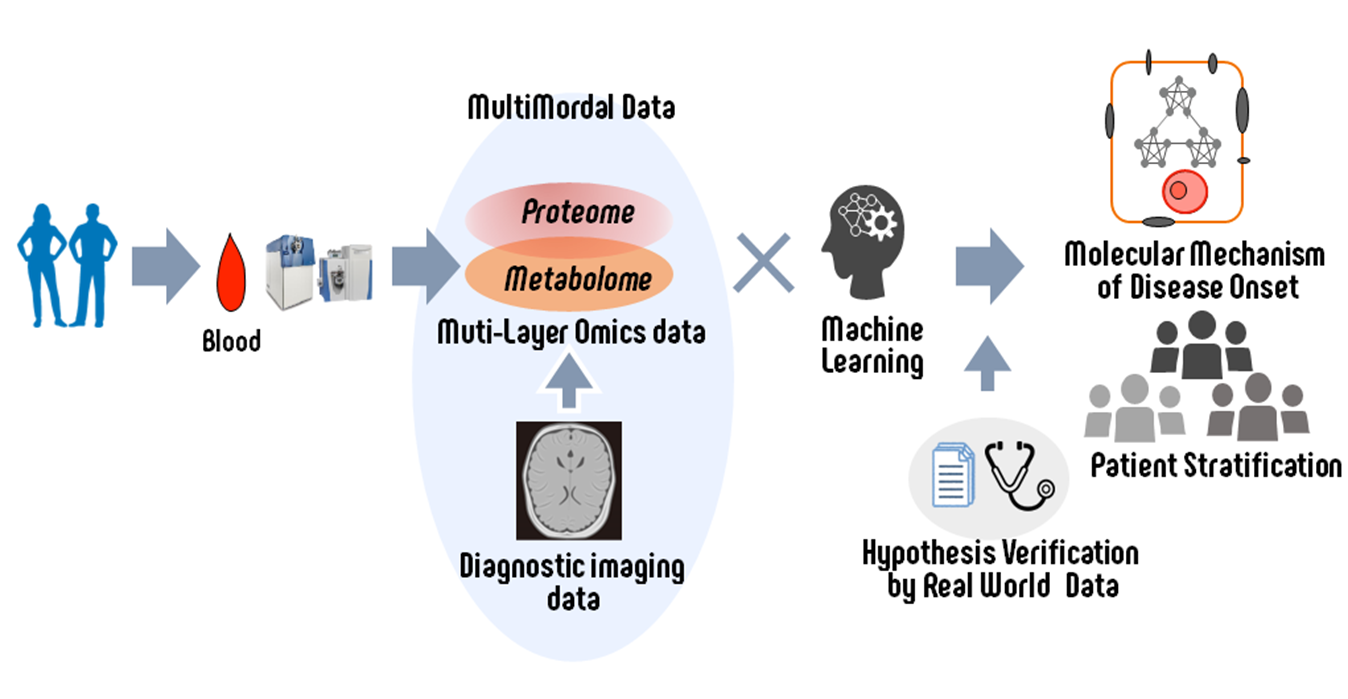
研究課題1:マルチモーダルデータ解析による疾患メカニズム推定及び診断のためのデータサイエンス
研究担当者:今井 賢一郎(グループ内連携:鍵和田 晴美・今野 雅允)
臨床検体の多層オミックスデータに診断画像等の解析データも組み合わせ、マルチモーダルなデータ解析を行い、疾患発症に関わる分子メカニズムの推定や早期診断に繋がる技術開発に取り組んでいます。また、レセプトデータなどのリアルワールドデータを活用し、得られた発症メカニズムの検証にも取り組んでいます。
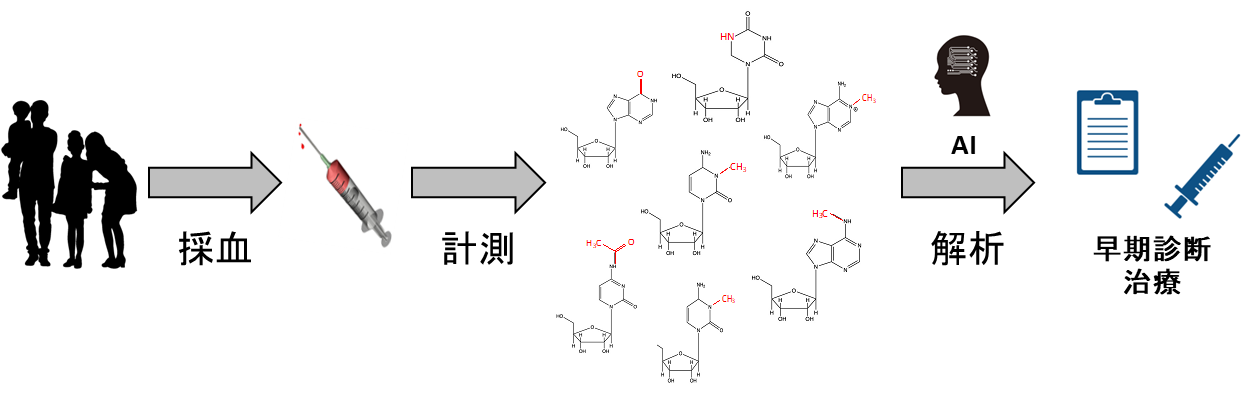
研究課題2:オミクス修飾を利用した疾患新規診断、治療法の開発
研究担当者:今野 雅允(グループ内連携者:小関 準・今井 賢一郎)
RNA修飾は様々な疾患で変化していることが知られています。しかその計測技術が未熟であり、診断治療への応用は実現していません。そこで私たちは、RNAを中心としたオミクスの修飾情報を計測する新規技術の開発を進めています。またAI等を用いて計測したオミクスの修飾情報解析を進め、がんをはじめとした様々な疾患の早期診断技術開発、新規治療法開発に取り組んでいます。
研究課題3:理論構造解析に基づく創薬支援技術の開発
研究担当者:小関 準(グループ内連携:本野 千恵・今井 賢一郎)
タンパク質の『構造』が変わると、その『機能』にも影響を与えることが知られています。しかしながら構造がわかっていないタンパク質については、その関係性をうまく捉えることができませんでした。最近ではAIを使用し、タンパク質の立体構造を予測できるようになってきましたが、どこが重要な部分なのか、またアミノ酸変異によってどのように働きが変わるのかを予測するのは困難です。そこで私たちは、この問題を解決するための理論構造解析法を開発し、Undruggableなタンパク質の隠れた機能部位を探索することで、低分子から中分子、さらには結合タンパク質を設計することで、創薬支援を目指しています。
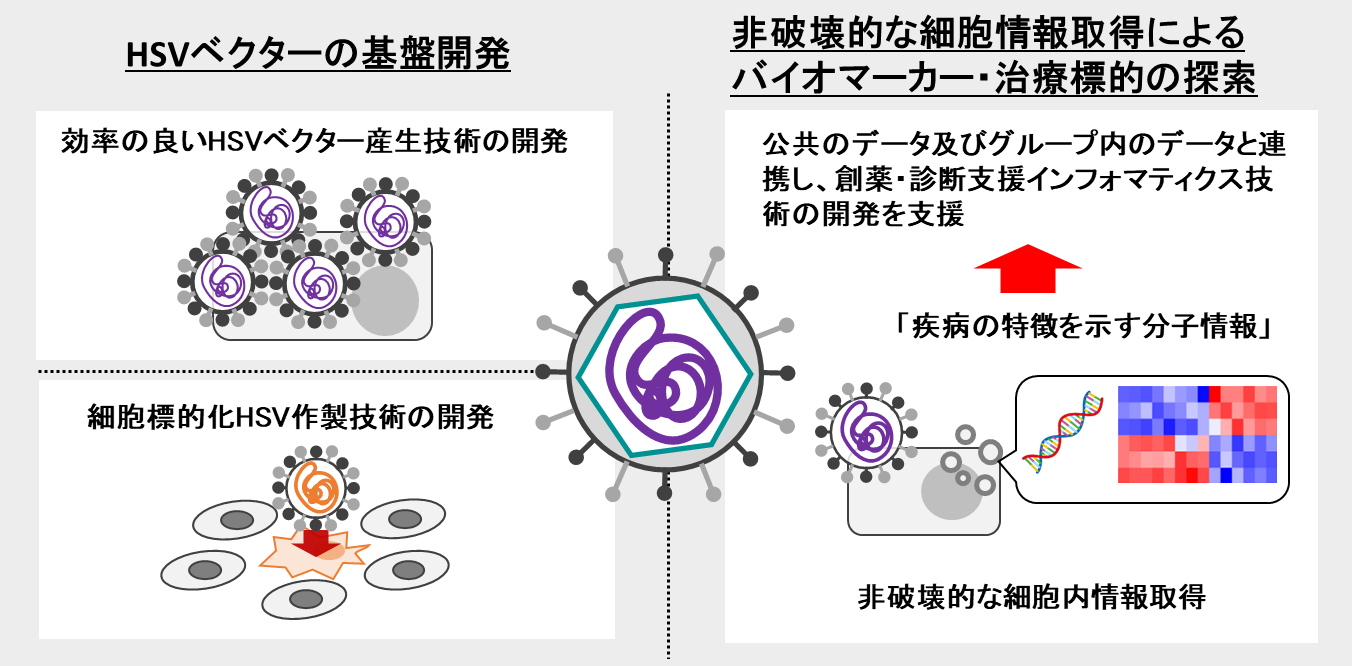
研究課題4:ヘルペスウイルスベクターの医薬モダリティとしての開発
研究担当者:前田 史雄(グループ内連携:小関 準・今井 賢一郎)
単純ヘルペスウイルスAmpliconベクターは、ウイルス粒子からウイルスゲノムを排除し、代わりにプラスミドをウイルス粒子にパッケージングしており、(1)極めて低い細胞毒性、(2)最大150kbpもの遺伝子積載容量、(3)幅広い種類の細胞への感染能を特徴としたウイルスベクターです。 現在、効率的なベクター産生法の開発、特定の細胞を標的化した遺伝子導入技術の開発、更にヘルペスウイルスベクターを遺伝子疾患に対する治療薬として利用する開発に取り組んでいます。
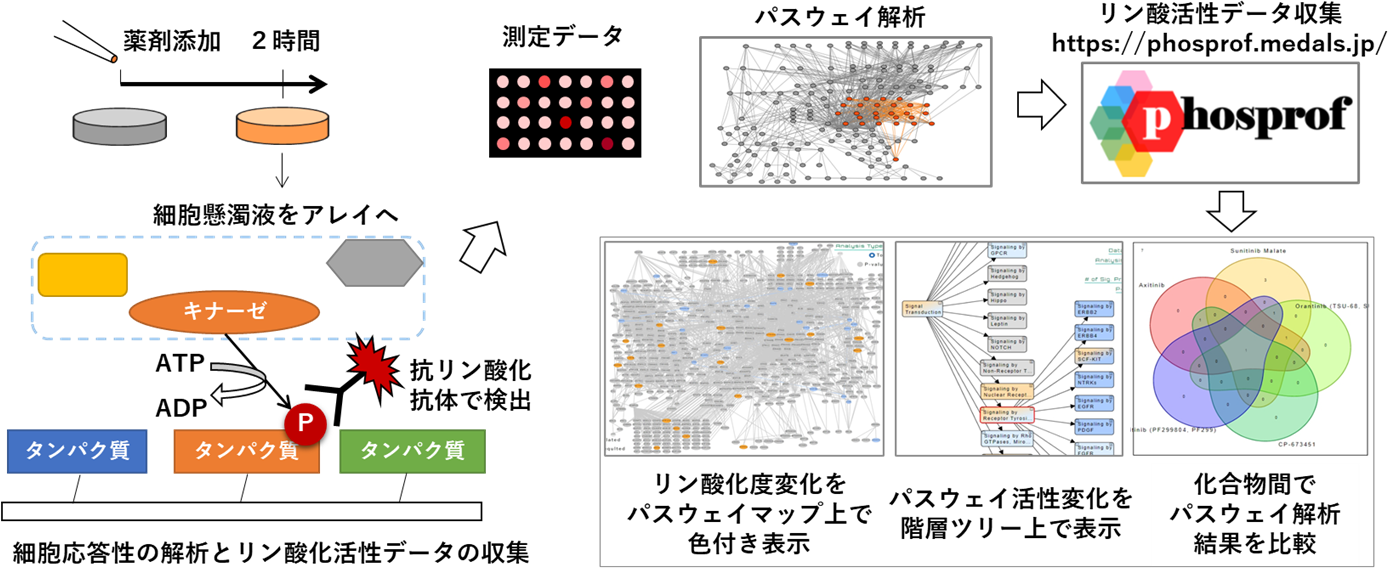
研究課題5:タンパク質群ネットワーク動態の解析基盤の構築
研究担当者:鍵和田 晴美(グループ内連携:前田 史雄・今井 賢一郎)
細胞外部からの刺激(シグナル)の多くはリン酸化経路によって伝えられ、細胞の挙動を決定しています。このシグナル伝達に関わる様々なタンパク質へのリン酸化活性を、たんぱく質アレイを使って一斉に測定し、パスウェイ解析を行う新しい評価系を開発しました。また、この系を用いて新たに94種類の市販薬を投与した細胞でリン酸化活性測定とパスウェイ解析を行った結果を収集し、データベースとして公開しています。現在、細胞・組織・分泌物などさまざまな解析対象でリン酸化活性の解析を試みています。
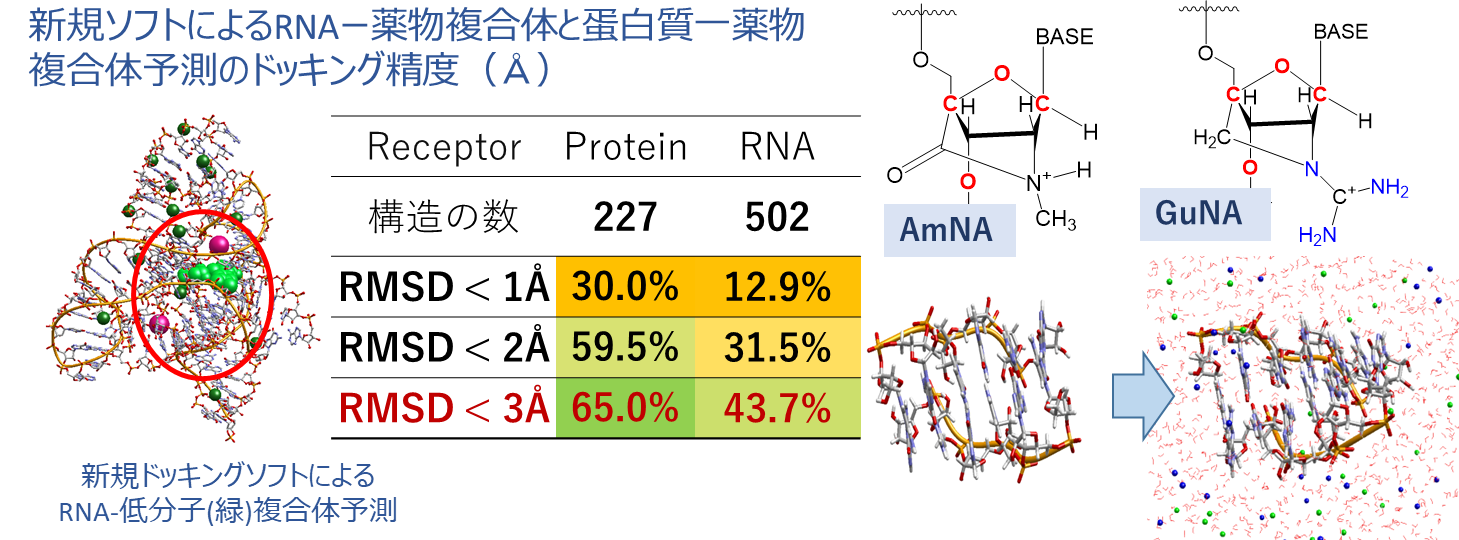
研究課題6:新規医薬品・医療技術のためのRNA分子シミュレーションの基礎技術開発
研究担当者:福西 快文(グループ内連携:本野 千恵・今井 賢一郎)
新型コロナワクチン以来、医薬分子(モダリティ)・医薬標的・検査マーカとしてRNAが拡大していますが、多様な副反応が示すように未知の性質が多く、本格的な普及は5~10年後と思われます。特にRNAは配列情報・立体構造に加え、分子運動により機能を発揮しますが、実験の困難さから構造情報は蛋白質の1/300しかありません。大量のデータを学習して予測力を発揮するAIにとって扱いにくいテーマです。我々は、核酸の理論計算精度を実験と比較する手法を開発し、計算法の最適化を図っています。私たちは、国内40社以上の製薬・IT企業と、共同で培ってきた計算技術を創薬ソフトウェア群myPrestoに実装し、生命活動を調整する生体分子の運動と分子認識機構、塩基の修飾などの制御機構を解明する理論的ツールの開発と提供を行います。
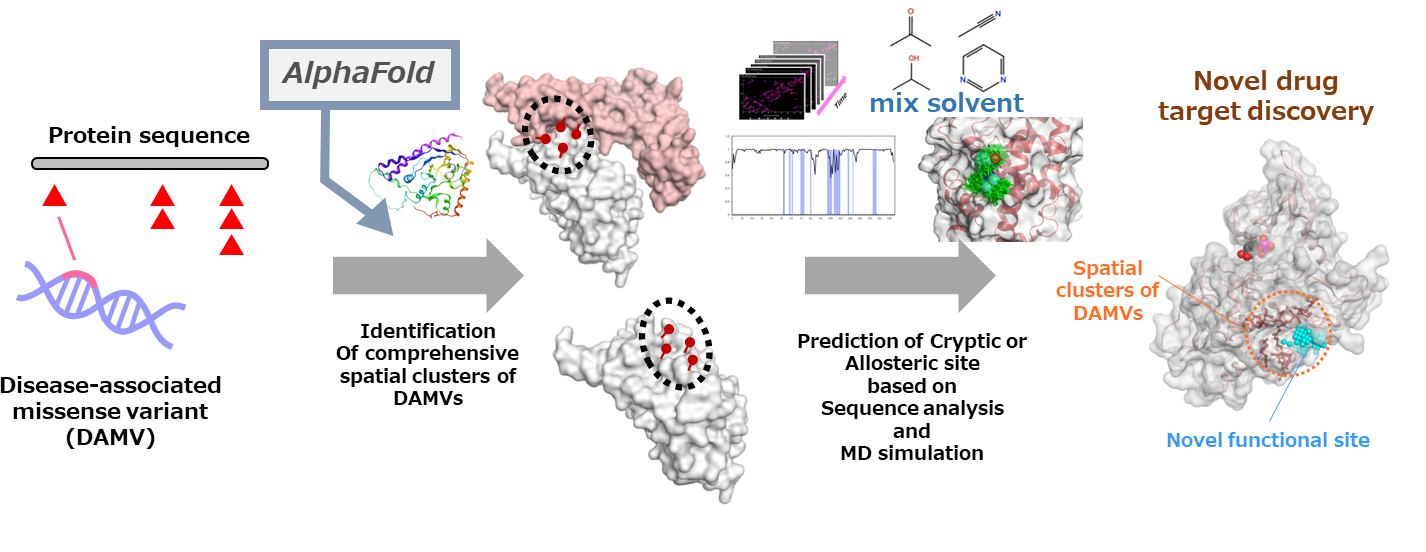
研究課題7:疾患関連変異情報を利用した新規創薬標的探索法の構築
研究担当者:本野 千恵 (グループ内連携:鍵和田 晴美・小関 準・今井 賢一郎)
近年、多くの疾患関連変異情報が得られていますが、それらの多くは、生物学的解釈が難しく、従来とは異なる視点での利活用法や解析技術が求められています。本研究では、疾患関連変異をタンパク質の立体構造上でのクラスタという単位でとらえ、それらを基点としたタンパク質の未発見の機能部位の網羅的探索法を開発しています。疾患関連変異は、タンパク質構造上で機能部位近辺に集積する傾向がある一方、既知の機能部位に紐づかないクラスタは、未発見の機能部位を指し示す可能性があります。また、未発見の機能制御部位は、単純なポケットではなく、動的に生じるリガンド結合部位(クリプトサイト)やアロステリック制御部位の可能性があります。そこで、配列解析や分子動力学計算などを組み合わせることで、新規の機能部位、ひいては、新規創薬標的部位を予測する技術開発を行っています。
グループの構成メンバー
| 顔写真 | 所属・役職および名前 | 専門分野 | その他、etc |
|---|---|---|---|
 |
研究グループ長(兼務) 今井 賢一郎 |
|
|
 |
主任研究員 今野 雅允 |
|
|
 |
主任研究員 小関 準 |
|
|
 |
主任研究員 鍵和田 晴美 |
|
|
 |
研究員 前田 史雄 |
|
|
 |
研究グループ付(兼務) 福西 快文 |
|
|
 |
研究グループ付(兼務) 本野 千恵 |
|
業績リスト
- Bekker, GJ; Fukunishi, Y; Higo, J; Kamiya, N.
Assessment of RNA Force Fields for Dynamic Docking of Small Molecules Using Multicanonical MD Simulations.
JOURNAL OF CHEMICAL THEORY AND COMPUTATION. 2025 Oct 22. doi:10.1021/acs.jctc.5c01114 - Ojima-Kato, T; Yokoyama, G; Nakano, H; Hamada, M; Motono, C.
Screening and machine learning-based prediction of translation-enhancing peptides that reduce ribosomal stalling in Escherichia coli.
RSC CHEM BIOL. 2025 Oct 22. doi:10.1039/d5cb00199d - Hara, T; Meng, S; Motooka, D; Arao, Y; Saito, Yk Rennie, S; Uchida, S; Ofusa, K; Arai, T; Konno, M; Satoh, T; Ishii, H.
Deficiencies in methionine, tryptophan, and niacin remodels intestinal transcriptome and gut microbiota in female mice.
SCI REP. 2025 Oct 16;15(1):36155. doi:10.1038/s41598-025-18046-2 - Lintuluoto, M; Horioka, Y; Fujiwara, M; Abe, M; Fukunishi, Y; Tamura, H; Lintuluoto, JM.
Investigation on the Substrate Specificity of Serine Protease Neuropsin by Molecular Dynamics Simulation and Marmoset Gene Atlas (MGA.
ACS OMEG. 2025 JUN 2. doi:10.1021/acsomega.5c00843 - Motono, C; Yanagisawa, K; Koseki, J; Imai, K.
CrypTothML: An Integrated Mixed-Solvent Molecular Dynamics Simulation and Machine Learning Approach for Cryptic Site Prediction.
INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2025 May 14;26(10):4710. doi:10.3390/ijms26104710 - Koseki, J; Motono, C; Yanagisawa, K; Kudo, G; Yoshino, R; Hirokawa, T; Imai, K.
CrypToth: Cryptic Pocket Detection through Mixed-Solvent Molecular Dynamics Simulations-Based Topological Data Analysis.
JOURNAL OF CHEMICAL INFORMATION AND MODELING. 2025 May 22. doi:10.1021/acs.jcim.4c02111 - Nobe, M; Maruzuru, Y; Takeshima, K; Maeda, F; Kusano, H; Yoshimura, R; Nishiyama, T; Park, H; Kozaki, Y; Iwami, S; Koyanagi, N; Kato, A; Natsume, T; Adachi, S; Kawaguchi, Y.
Direct relationship between protein expression and progeny yield of herpes simplex virus 1.
MBIO. 2025 May 5:e0028025. doi:10.1128/mbio.00280-25 - Ohkawa, M; Ohshiro, T; Koseki, J; Kon, S; Ozeki, Y; Taniguchi, M; Shimamura, T; Konno, M.
New RNA modifications induce malignancy for pancreatic cancer.
CANCER SCIENCE. 2025 Mar. doi: 10.1111/cas.16114 - Yamagishi, A; Tokuoka, R; Imai, K; Mizusawa, M; Susaki, M; Uchida, K; Kijima, ST; Nagasaki, A; Takeshita, D; Yoshikawa, C; Uyeda, TQP; Nakamura, C.
Nestin Forms a Flexible Cytoskeleton by Means of a Huge Tail Domain That Is Reversibly Stretched and Contracted by Weak Forces.
CELLS. 2025 Jan 17;14(2):138. doi:10.3390/cells14020138 - Sato, T; Abe, K; Koseki, J; Seto, M; Yokoyama, J; Akashi, T; Terada, M; Kadowaki, K; Yoshida, S; Yamashiki, YA; Shimamura, T.
Survivability and life support in sealed mini-ecosystems with simulated planetary soils.
SCI REP. 2024 Nov 1;14(1):26322. doi: 10.1038/s41598-024-75328-x - Yoshinori, F; Imai, K; Horton, P.
Prediction of mitochondrial targeting signals and their cleavage sites.
METHODS ENZYMOL. 2024 Sep 11. doi: 10.1016/bs.mie.2024.07.026 - Fujii, Y; Kamata, K; Gerdol, M; Hasan, I; Rajia, S; Kawsar, SMA; Padma, S; Chatterjee, BP; Ohkawa, M; Ishiwata, R; Yoshimoto, S; Yamada, M; Matsuzaki, N; Yamamoto, K; Niimi, Y; Miyanishi, N; Konno, M; Pallavicini, A; Kawasaki, T; Ogawa, Y; Ozeki, Y; Fujita, H.
Multifunctional Cell Regulation Activities of the Mussel Lectin SeviL: Induction of Macrophage Polarization toward the M1 Functional Phenotype.
MARINE DRUGS. 2024 Jun 11;22(6):269. doi: 10.3390/md22060269 - Watanabe, A; Koyano, F; Imai, K; Hizukuri, Y; Ogiwara, S; Ito, T; Miyamoto, J; Shibuya, C; Kimura, M; Toriumi, K; Motono, C; Arai, M; Tanaka, K; Akiyama, Y; Yamano, K; Matsuda, N.
The origin of esterase activity of Parkinsons disease causative factor DJ-1 implied by evolutionary trace analysis of its prokaryotic homolog HchA.
J BIOL CHEM. 2024 Jun 13:107476. doi: 10.1016/j.jbc.2024.107476 - Higo, J; Bekker, GJ; Kamiya, N; Fukuda, I; Fukunishi, Y;.
Binding free-energy landscapes of small molecule binder and non-binder to FMN riboswitch: All-atom molecular dynamics.
BIOPHYS PHYSICOBIOL. 2023 Dec 13;20(4):e200047. doi: 10.2142/biophysico.bppb-v20.0047 - Bekker, GJ; Fukunishi, Y; Higo, J; Kamiya, N.
Binding Mechanism of Riboswitch to Natural Ligand Elucidated by McMD-Based Dynamic Docking Simulations.
ACS OMEGA. 2024 Jan 10;9(3):3412-3422. doi: 10.1021/acsomega.3c06826 eCollection - Germany, EM; Thewasano, N; Imai, K; Maruno, Y; Bamert, RS; Stubenrauch, CJ; Dunstan, RA; Ding, Y; Nakajima, Y; Lai, X; Webb, CT; Hidaka, K; Tan, KS; Shen, H; Lithgow, T; Shiota, T.
Dual recognition of multiple signals in bacterial outer membrane proteins enhances assembly and maintains membrane integrity.
ELIFE. 2024 Jan 16;12:RP90274. doi: 10.7554/eLife.90274 - Shimizu, R; Murai, K; Tanaka, K; Sato, Y; Takeda, N; Nakasyo, S; Shirasaki, T; Kawaguchi, K; Shimakami, T; Nio, K; Nakaya, Y; Kagiwada, H Horimoto, K; Mizokami, M; Kaneko, S; Murata, K; Yamashita, T; Honda, M.
Nucleos(t)ide analogs for hepatitis B virus infection differentially regulate the growth factor signaling in hepatocytes.
HEPATOL COMMUN. 2024 Jan 5;8(1):e0351. doi: 10.1097/HC9.0000000000000351 - Fukuda, I; Moritsugu, K; Higo, J; Fukunishi, Y.
A cutoff-based method with charge-distribution-data driven pair potentials for efficiently estimating electrostatic interactions in molecular systems.
J CHEM PHYS. 2023 Dec 21;159(23):234116. doi: 10.1063/5.0172270 - Watanabe, Y; Iwasaki, Y; Sasaki, K; Motono, C; Imai, K; Suzuki, K.
Atg15 is a vacuolar phospholipase that disintegrates organelle membranes.
CELL REP. 2023 Dec 15:113567. doi: 10.1016/j.celrep.2023.113567 - Maeda, F; Adachi, S; Natsume, T.
Non-destructive and efficient method for obtaining miRNA information in cells by artificial extracellular vesicles.
SCI REP. 2023 Dec 14;13(1):22231. doi: 10.1038/s41598-023-48995-5 - Ohkawa, M; Konno, M.
RNA Modification Related Diseases and Sensing Methods.
APPLIED SCIENCES-BASEL. 2023 13(11) 6376. doi: 10.3390/app13116376 - Shin, WH; Kumazawa, K; Imai, K; Hirokawa, T; Kihara, D.
Quantitative comparison of protein-protein interaction interface using physicochemical feature-based descriptors of surface patches.
FRONT MOL BIOSCI. 2023 Feb 6;10:1110567. doi: 10.3389/fmolb.2023.1110567 - Takeda, H; Busto, JV; Lindau, C; Tsutsumi, A; Tomii, K; Imai, K; Yamamori, Y; Hirokawa, T; Motono, C; Ganesan, I; Wenz, LS; Becker, T; Kikkawa, M; Pfanner, N; Wiedemann, N; Endo, T.
A multipoint guidance mechanism for β-barrel folding on the SAM complex.
NAT STRUCT MOL BIOL. 2023 Jan 5. doi: 10.1038/s41594-022-00897-2 - Hayashida, R; Kikuchi, R; Imai, K; Kojima, W; Yamada, T; Iijima, M; Sesaki, H; Tanaka, K; Matsuda, N; Yamano, K.
Elucidation of ubiquitin-conjugating enzymes that interact with RBR-type ubiquitin ligases using a liquid-liquid phase separation-based method.
J BIOL CHEM. 2022 Dec 20:102822. doi: 10.1016/j.jbc.2022.102822 - Murotomi, K; Kagiwada, H; Hirano, K; Yamamoto, S; Numata, N; Matsumoto, Y; Kaneko, H; Namihira, M.
Cyclo-glycylproline attenuates hydrogen peroxide-induced cellular damage mediated by the MDM2-p53 pathway in human neural stem cells.
J CELL PHYSIOL. 2022 Dec 31. doi: 10.1002/jcp.30940 - Fukunishi, Y; Higo, J; Kasahara, K.
Computer simulation of molecular recognition in biomolecular system: from in silico screening to generalized ensembles.
BIOPHYS REV. 2022 Nov 28:1-25. doi: 10.1007/s12551-022-01015-8 - Lintuluoto, M; Abe, M; Horioka, Y; Fukunishi, Y; Tamura, H; M, Lintuluoto J.
Investigation on substrate specificity and catalytic activity of serine protease neuropsin.
BIOPHYS PHYSICOBIOL. 2022 Sep 22;19:e190040. doi: 10.2142/biophysico.bppb-v19.0040 - Kagiwada, H; Motono, C; Horimoto, K; Fukui, K.
Phosprof: pathway analysis database of drug response based on phosphorylation activity measurements.
DATABASE (Oxford). 2022 Aug 22;2022:baac072. doi: 10.1093/database/baac072 - Higo, J; Kasahara, K; Bekker, GJ; Ma, B; Sakuraba, S; Iida, S; Kamiya, N; Fukuda, I; Kono, H; Fukunishi, Y; Nakamura, H.
Fly casting with ligand sliding and orientational selection supporting complex formation of a GPCR and a middle sized flexible molecule.
SCI REP. 2022 Aug 13;12(1):13792. doi: 10.1038/s41598-022-17920-7 - Hirata, Y; Oda, AH; Motono, C; Shiro, M; Ohta, K.
Imputation-free reconstructions of three-dimensional chromosome architectures in human diploid single-cells using allele-specified contacts.
SCI REP. 2022 Jul 11;12(1):11757. doi: 10.1038/s41598-022-15038-4 - Kose, S; Imai, K; Watanabe, A; Nakai, A; Suzuki, Y; Imamoto, N.
Lack of Hikeshi activates HSF1 activity under normal conditions and disturbs the heat-shock response.
LIFE SCI ALLIANCE. 2022 May 17;5(9):e202101241. doi: 10.26508/lsa.202101241 - Maeda, F; Kato, A; Takeshima, K; Shibazaki, M; Sato, R; Shibata,T; Miyake, K; Kozuka-Hata, H; Oyama, M; Shimizu, E; Imoto, S; Miyano, S; Adachi, S; Natsume, T; Takeuchi, K; Maruzuru, Y; Koyanagi, N; Jun, A; Yasushi, K.
Role of the Orphan Transporter SLC35E1 in the Nuclear Egress of Herpes Simplex Virus 1.
J VIROL. 2022 Apr 27:e0030622. doi: 10.1128/jvi.00306-22 - Santos, HJ; Hanadate, Y; Imai, K; Watanabe, H; Nozaki, T.
Entamoeba histolytica EHD1 Is Involved in Mitosome-Endosome Contact.
MBIO. 2022 Apr 11:e0384921. doi: 10.1128/mbio.03849-21 - Araiso, Y; Imai, K; Endo, T.
Role of the TOM Complex in Protein Import into Mitochondria: Structural Views.
ANNU REV BIOCHEM. 2022 Feb 14. doi: 10.1146/annurev-biochem-032620-104527 - Ono, J; Koshimizu, U; Fukunishi, Y; Nakai, H.
Multiple protonation states in ligand-free SARS-CoV-2 main protease revealed by large-scale quantum molecular dynamics simulations.
CHEM PHYS LETT. 2022 Feb 22;794:139489. doi: 10.1016/j.cplett.2022.139489 - Queliconi BB, Kojima W, Kimura M, Imai K, Udagawa C, Motono C, Hirokawa T, Tashiro S, Caaveiro JMM, Tsumoto K, Yamano K, Tanaka K, Matsuda N.
Unfolding is the driving force for mitochondrial import and degradation of Parkinsons disease-related protein DJ-1.
J CELL SCI. 2021 Oct 22:jcs.258653. doi: 10.1242/jcs.258653 - Kimura, M; Imai, K; Morinaka, Y; Hosono-Sakuma, Y; Horton, P; Imamoto, N.
Distinct mutations in importin-β family nucleocytoplasmic transport receptors transportin-SR and importin-13 affect specific cargo binding.
SCI REP. 2021 Aug 2;11(1):15649. doi: 10.1038/s41598-021-94948-1 - Motono, C; Yanagida, S; Sato, M; Hirokawa, T.
MDContactCom: a tool to identify differences of protein molecular dynamics from two MD simulation trajectories in terms of interresidue contacts.
BIOINFORMATICS. 2021 Jul 21:btab538. doi: 10.1093/bioinformatics/btab538 - Iida, S; Fukunishi, Y.
Asymmetric dynamics of dimeric SARS-CoV-2 and SARS-CoV main proteases in an apo form: Molecular dynamics study on fluctuations of active site, catalytic dyad, and hydration water.
BBA ADV. 2021;1:100016. doi: 10.1016/j.bbadva.2021.100016. Epub 2021 Jun 20 - Kagiwada, H; Kiboku, T; Matsuo, H; Kitazawa, M; Fukui, K; Horimoto, K.
Assessing the activation/inhibition of tyrosine kinase-related pathways with a newly developed platform.
PROTEOMICS. 2021 Jun 21:e2000251. doi: 10.1002/pmic.202000251 - Moritsugu, K; Takeuchi, K; Kamiya, N; Higo, J; Yasumatsu, I; Fukunishi, Y; Fukuda, I.
Flexibility and Cell Permeability of Cyclic Ras-Inhibitor Peptides Revealed by the Coupled Nosé-Hoover Equation.
J CHEM INF MODEL. 2021 Apr 9. doi: 10.1021/acs.jcim.0c01427 - Lintuluoto, M; Horioka, Y; Hongo, S; Lintuluoto, JM; Fukunishi, Y.
Molecular Dynamics Simulation Study on Allosteric Regulation of CD44-Hyaluronan Binding as a Force Sensing Mechanism.
ACS OMEGA. 2021 Mar 16;6(12):8045-8055. doi: 10.1021/acsomega.0c05502. eCollection