生合成システム多様性研究グループ
研究概要
生合成システム多様性研究グループのミッションは、生物が持つ能力と化学の知識を用いて社会課題の解決に取り組むことです。微生物、植物、動物における生命現象の解明を行い、学問の発展に寄与するとともに、これらの理解に基づいた物質生産や創薬に資する生物資源と技術を創出し、バイオエコノミー社会の実現に貢献することを目指します。

研究課題
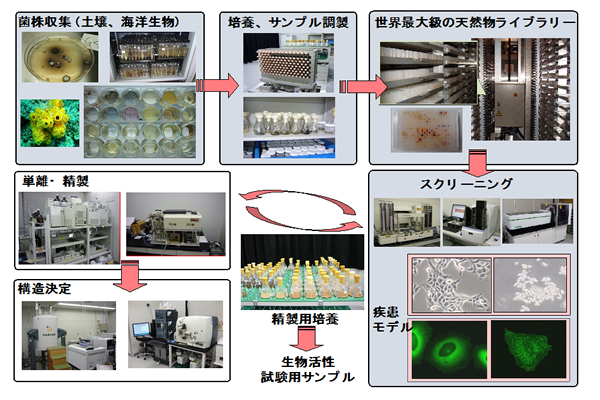
研究課題1:世界最大級の天然物ライブラリーを用いた創薬スクリーニング
研究担当者:工藤 慧、坂本 良太
現在上市されている医薬品の6割以上は天然化合物あるいは天然化合物を模倣して合成展開した化合物に由来するように、天然物ライブラリーは創薬スクリーニングのソースとして長く用いられています。我々は、国内有数の製薬系企業提供の天然物ライブラリーを含む、世界最大級の天然物ライブラリーを保有しています。本天然物ライブラリーを用いて、企業およびアカデミア発の創薬ターゲットを対象に、大規模な医薬、動物薬、農薬等のリード化合物探索を展開しています。
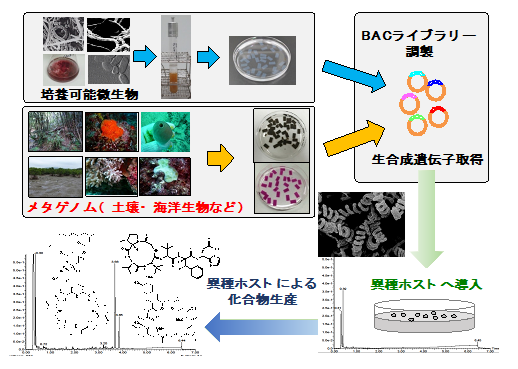
研究課題2:生合成遺伝子を用いた異種発現システムによる天然化合物生産技術開発
研究担当者:工藤 慧、Pisanee Srisawat、坂本 良太
従来の発酵法による微生物二次代謝生産では、微生物ゲノム中に存在する生合成遺伝子数の1/3にも満たない能力しか引き出せていないことが明らかになってきています。本研究開発では、難培養微生物由来の未利用生合成遺伝子を含め、種々の微生物の生合成遺伝子を異種ホストに形質転換し、異種発現システムにより、これまで人類が手に入れることの出来なかった新奇天然化合物を創出することを目的としています。また、世界で我々だけが持つ複雑な構造からなる天然化合物の母骨格改変技術を用いて、臨床薬開発に必須な化合物ファインチューニングに貢献します。
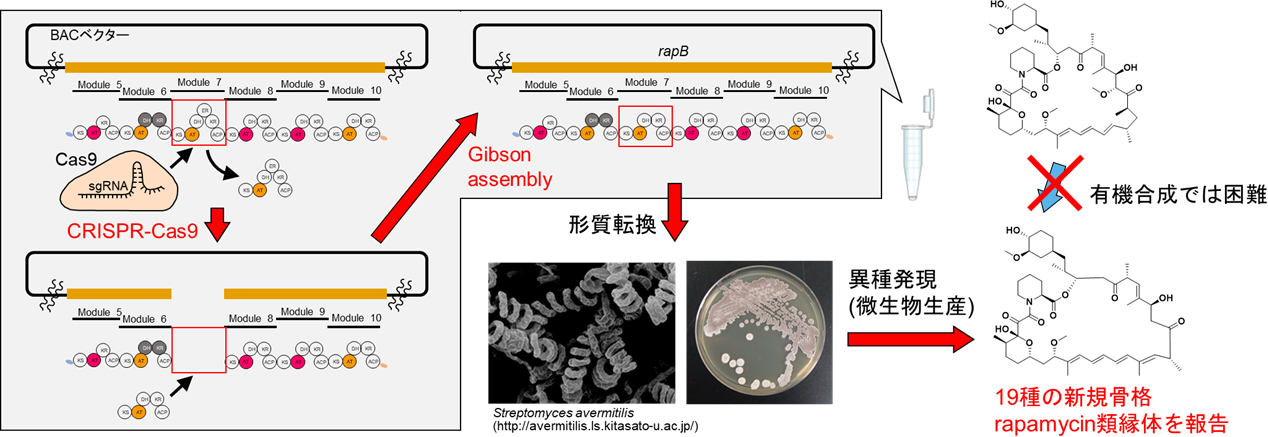
研究課題3:超精密合成遺伝子改変技術による誘導体展開
研究担当者:工藤 慧、Pisanee Srisawat
天然化合物は医薬品等に応用される生物活性物質の重要なリソースです。極めて複雑な構造を有する天然化合物の誘導体展開は、現代の有機合成でも容易ではなく、新しい手法が求められています。近年我々は、「ポリケタイド」と呼ばれる化合物群の生合成遺伝子を超精密に改変する技術(in vitroモジュール編集技術)の開発に成功しました。ポリケタイド合成酵素遺伝子は非常に類似性が高い配列を繰り返し有するため、従来の遺伝子組換え技術では正確な改変は不可能でしたが、我々の技術を用いることで、自由自在に遺伝子を挿入・置換・欠失させ、新規ポリケタイドを創出することが可能となりました。本技術を基盤として、ポリケタイド骨格の多様性を飛躍的に増大させる技術(コンビナトリアル生合成技術)の開発や、天然化合物と新しい創薬モダリティとの融合、バイオものづくり産業に貢献する新しい有用物質の創出など、基礎及び応用研究を展開しています。
研究課題4:RNAの修飾塩基の機能と生合成の多様性の解明と応用展開
研究担当者:鴫 直樹
ゲノム機能が的確に発揮されるためには、タンパク質の翻訳後修飾やRNAの転写後修飾が重要であることが近年注目を集めています。RNAが正確に機能するためには、複雑なRNA修飾を受けることが重要である。そこでRNA修飾経路の多様性とその生体内意義を研究します。RNA修飾の異常が疾患の原因となる例が近年多数報告されてきており、オリジナルな基礎研究から新しい創薬シーズの発見を目指します。また生物種による修飾機構の差異を明らかにすることから、副作用のない抗生物質の創出を目指します。
研究課題5:昆虫共生細菌の機能解明と応用技術基盤の開発
研究担当者:安佛 尚志
多くの昆虫の体内には様々な共生微生物が存在し、宿主の生理機能に多様な影響を与えています。例えば、昆虫の外骨格を硬くするために必須な共生細菌や、宿主昆虫の生殖様式を変えてしまう共生細菌が存在します。主に、シングルセルゲノム解析やモデル共生系を用いた実験により、その分子メカニズムの解明に取り組んでいます。昆虫は、害虫や益虫など人間社会との関係も深く、またショウジョウバエなどが疾病モデル生物としても用いられることから、昆虫個体群の制御や防除に資する基盤技術の開発や、創薬や治療を支援する技術の開発につながることが期待されます。
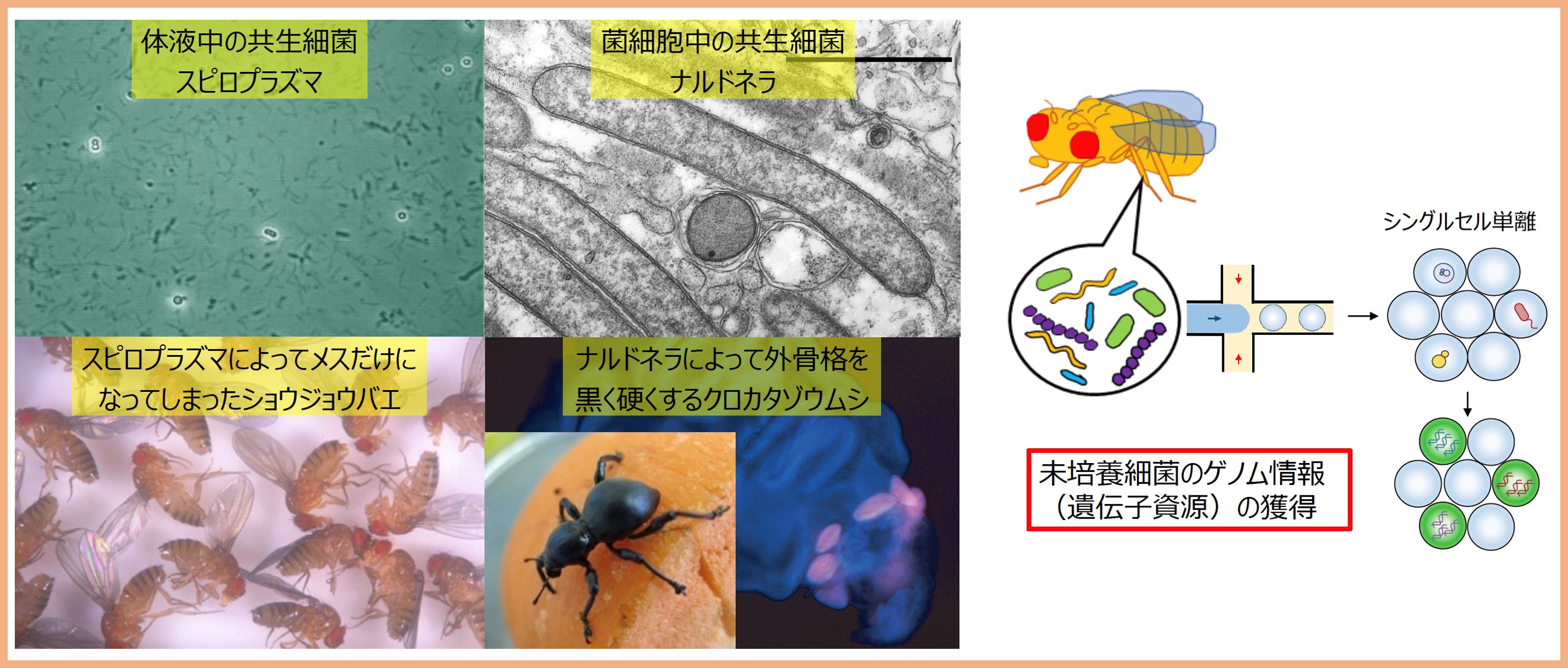
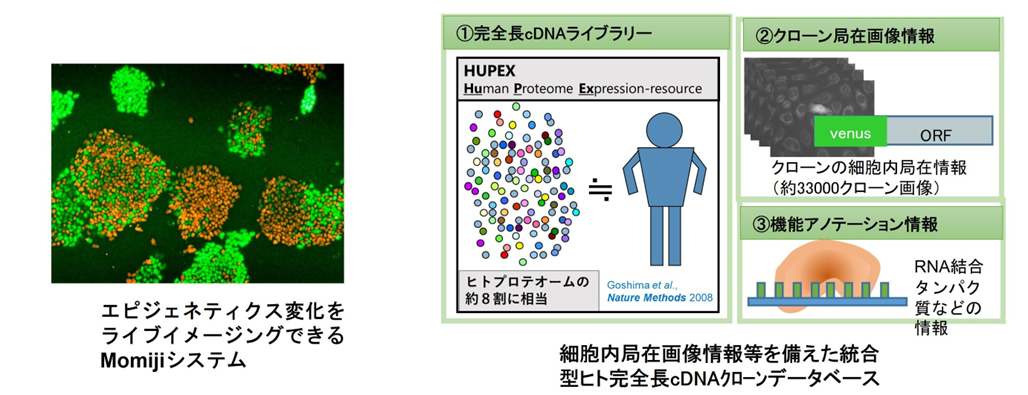
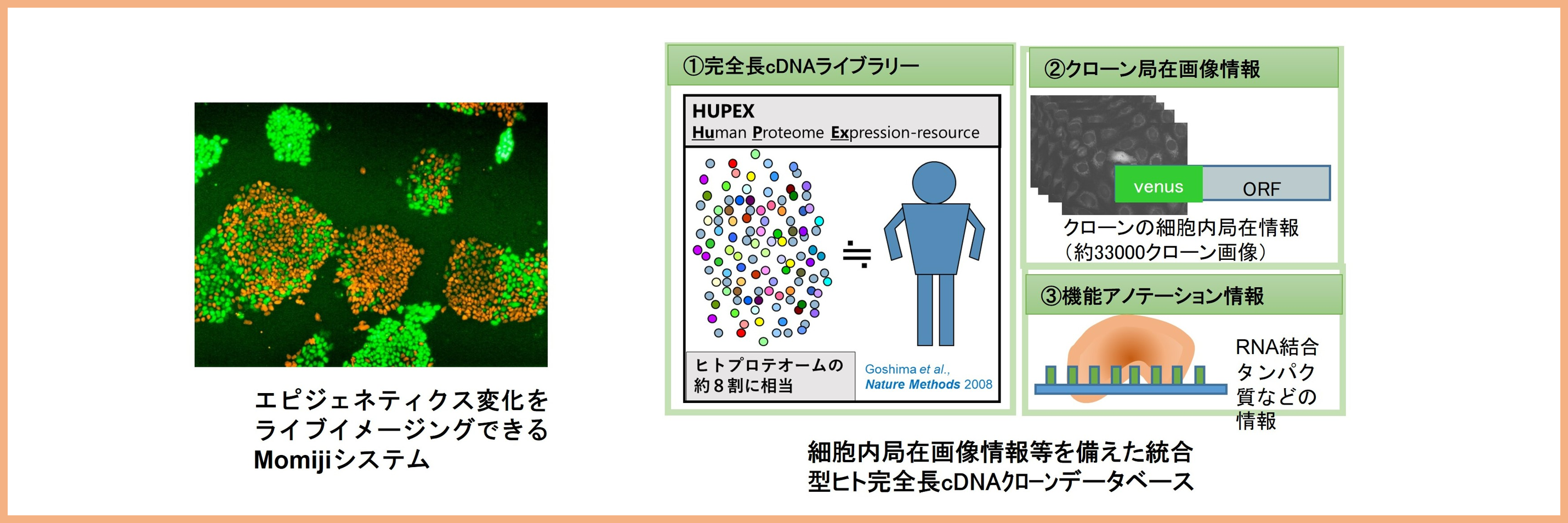
研究課題6:遺伝学的情報とは異なるアプローチによる細胞の理解を目指した解析基盤の確立
研究担当者:小林 慎
多くの昆虫の体内には様々な共生微生物が存在し、宿主の生理機能に多様な影響を与えています。例えば、昆虫の外骨格を硬くするために必須な共生細菌細胞内ではエピジェネティクスなど、単純な遺伝情報を超えた様々なイベントが起きています。我々はエピジェネティックな状態変化をライブイメージングする技術や、細胞内局在情画像報を備えた統合型ヒト完全長cDNAクローンデータベースなど活用し、遺伝学的情報を超えた細胞の動的な状態を研究し、生物の理解や創薬に繋げる研究を推進します。具体的には、哺乳類でみられるエピジェネティックな制御で知られる「X染色体不活性化」や、核内の様々な反応の制御・調節に働く「非膜構造体」などを対象とした研究を行っています。
グループの構成メンバー
| 顔写真 | 所属・役職および名前 | 専門分野 | その他、etc |
|---|---|---|---|
 |
グループ長 鴫 直樹 |
|
|
 |
主任研究員 安佛 尚志 |
|
|
 |
主任研究員 小林 慎 |
|
|
 |
主任研究員 工藤 慧 |
|
|
 |
研究員 Sriswat Pisanee |
|
|
 |
研究員 坂本 良太 |
|
業績リスト
- Mori, T; Zhou, MY; Kunugitani, K; Akatsuka, T; Yoshida, Y; Kouyama-Suzuki, E; Kobayashi, S; Shirai, Y; Tabuchi, K.
The Competitive Loss of Cerebellar Granule and Purkinje Cells Driven by X-Linked Mosaicism in a Female Mouse Model of CASK-Related Disorders.
CELLS. 2025 May 17;14(10):735. doi:10.3390/cells14100735 - Koshiguchi, M; Yonezawa, N; Hatano, Y; Suenaga, H; Yamagata, K; Kobayashi, S.
A system to analyze the initiation of random X-chromosome inactivation using time-lapse imaging of single cells.
SCI REP. 2024 Sep 2;14(1):20327. doi: 10.1038/s41598-024-71105-y - Nishimura, T; Kudo, K; Izumikawa, M; Kozone, I; Hashimoto, J; Kagaya, N; Suenaga, H; Takeuchi, K; Shin-Ya, K.
Isolation and Structure Elucidation of JBIR-157, a Skeletally Novel Aromatic Polyketide Produced by the Heterologous Expression of a Cryptic Gene Cluster.
CHEM PHARM BULL (Tokyo). 2024;72(5):475-479. doi: 10.1248/cpb.c24-00144 - Kudo, K; Nishimura, T; Izumikawa, M; Kozone, I; Hashimoto, J; Fujie, M; Suenaga, H; Ikeda, H; Satoh, N; Shin-Ya, K.
Capability of a large bacterial artificial chromosome clone harboring multiple biosynthetic gene clusters for the production of diverse compounds.
J ANTIBIOT (Tokyo). 2024 Mar 4. doi: 10.1038/s41429-024-00711-9 - Sugio, Y; Yamagami, R; Shigi, N; Hori, H.
A selective and sensitive detection system for 4-thiouridine modification in RNA.
RNA. 2022 Nov 21:rna.079445.122. doi: 10.1261/rna.079445.122 - Choirunnisa, AR; Arima, K; Abe,Y; Kagaya, K; Kudo, K; Suenaga, H; Hashimoto, J; Fujie, M; Satoh, N; Shin-Ya, K; Matsuda, K; Wakimoto, T.
New azodyrecins identified by a genome mining-directed reactivity-based screening.
BEILSTEIN J ORG CHEM. 2022 Aug 10;18:1017-1025. doi: 10.3762/bjoc.18.102 - Kojima, S; Shiochi, N; Sato, K; Yamaura, M; Ito, T; Yamamura, N; Goto, N; Odamoto, M; Kobayashi, S; Kimura, T; Sekita, Y.
Epigenome editing reveals core DNA methylation for imprinting control in the Dlk1-Dio3 imprinted domain.
NUCLEIC ACIDS RESEARCH. 2022 May 11. doi: 10.1093/nar/gkac344 - Aizawa, S; Nishimura, K; Mondejar, GS; Kumar, A; Bui, PL; Tran, YTH; Kuno, A; Muratani, M; Kobayashi, S; Nabekura, T; Shibuya, A; Sugihara, E; Sato, TA; Fukuda, A; Hayashi, Y; Hisatake, K.
Early reactivation of clustered genes on the inactive X chromosome during somatic cell reprogramming.
STEM CELL REPORTS. 2021 Nov 29:S2213-6711(21)00589-0. doi10.1016/j.stemcr.2021.11.008 - Zhang, M; Kiyono, T; Aoki, K; Goshima, N; Kobayashi, S; Hiranuma, K; Shiraishi, K; Saya, H; Nakahara, T.
Development of an in vitro carcinogenesis model of HPV induced cervical adenocarcinoma.
CANCER SCI. 2021 Dec 21. doi: 10.1111/cas.15246 - Shigi, N.
Biosynthesis and Degradation of Sulfur Modifications in tRNAs.
INT J MOL SCI. 2021 Nov 3;22(21):11937. doi: 10.3390/ijms222111937 - Kudo, K; Nishimura, T; Kozone, I; Hashimoto, J; Kagaya, N; Suenaga, H; Ikeda, H; Shin-Ya, K.
Hemiacetal-less rapamycin derivatives designed and produced by genetic engineering of a type I polyketide synthase.
SCI REP. 2021 May 11;11(1):9944. doi: 10.1038/s41598-021-88583-z - Liu, C; Hashimoto, J; Kudo, K; Shin-Ya, K; Kakeya, H.
An Atypical Arginine Dihydrolase Involved in the Biosynthesis of Cyclic Hexapeptide Longicatenamides.
CHEM ASIAN J. 2021 Apr 22. doi: 10.1002/asia.202100181 - Haramoto, Y; Sakata, M; Kobayashi, S.
Visualization of X chromosome reactivation in mouse primordial germ cells in vivo.
BIOL OPEN. 2021 Apr 15;10(4):bio058602. doi: 10.1242/bio.058602. Epub 2021 Apr 29