細胞機能デザイン研究グループ
研究概要
細胞機能デザイン研究グループでは、生物細胞が持つ物質生産能力を、高度にデザインし、細胞の性質を精密に制御・改変することで最大限に引き出し、最適化された細胞を創出することです。微生物・動物細胞の状態を把握する技術、細胞の性質を改変するための新規な設計原理を開発することで、物質生産や創薬に資する細胞を構築し、新たな細胞産業の創出に貢献することを目指します。
研究課題
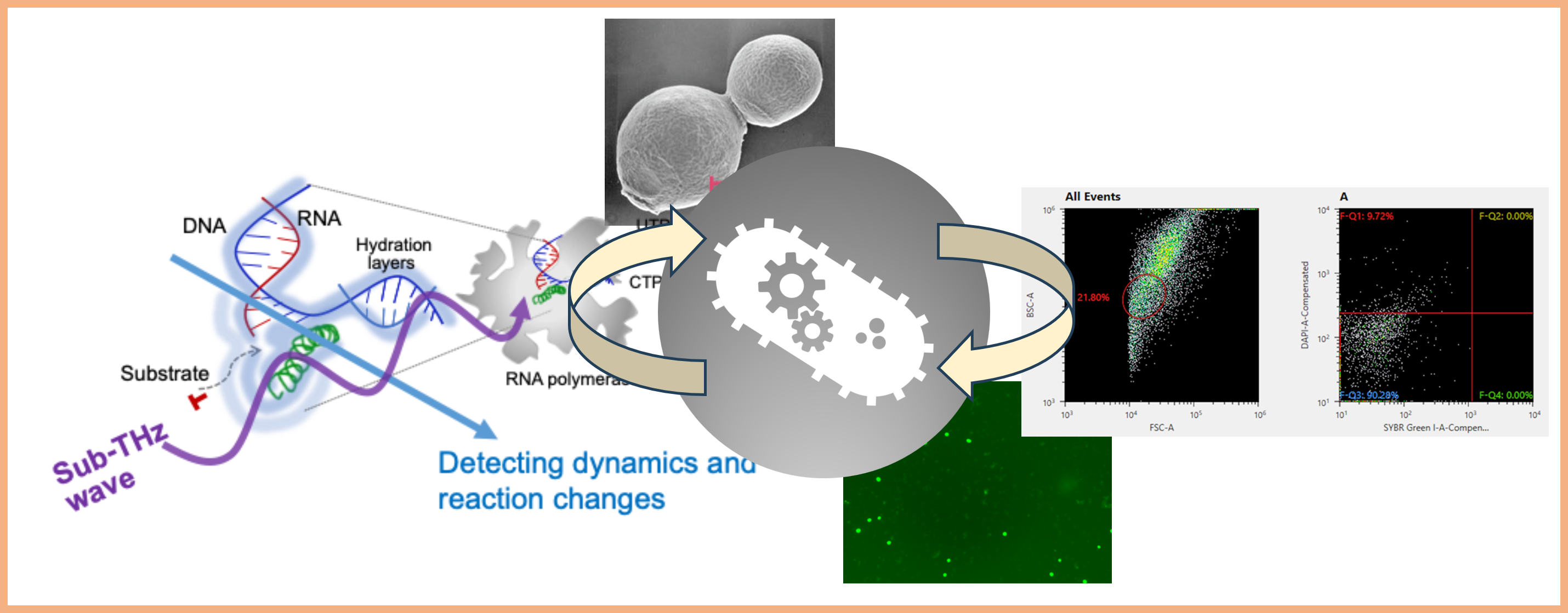
研究課題1:細胞を用いた有用物質生産
研究担当者:末永 光
微生物・動物細胞が持つ物質生産能力を、高度にデザインし、細胞の性質を精密に制御・改変することで最大限に引き出し、最適化された細胞を創出します。
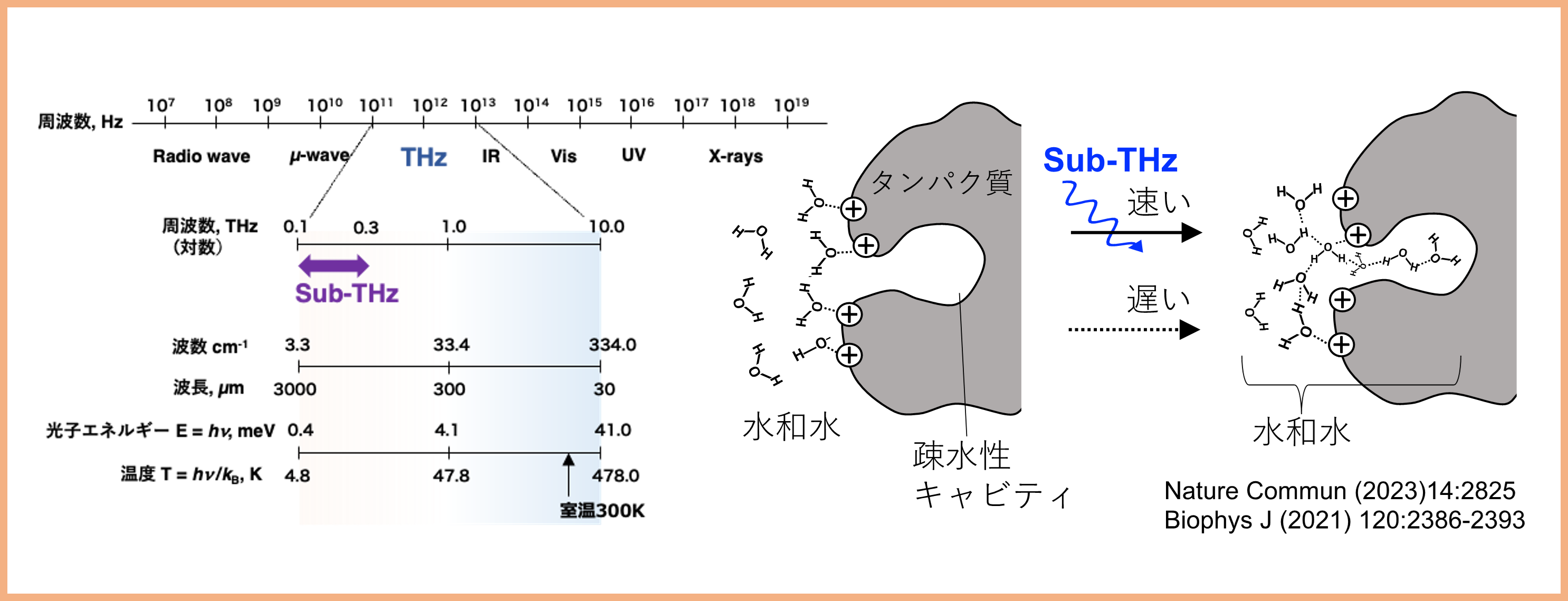
研究課題2:Sub-THz波を用いた細胞分子機能の制御、水和の制御
研究担当者:今清水 正彦
生物の精巧な分子機能の多くは、水と生体高分子の相互作用(その多くは水素結合)と、その揺らぎによって生み出されます。水和によって不均一化した水素結合の揺らぎは、THz周波数領域に現れます。「Sub-THz波による制御」の視点で、生物の分子機能の解明と複数分野に波及する応用展開に取り組んでいます。
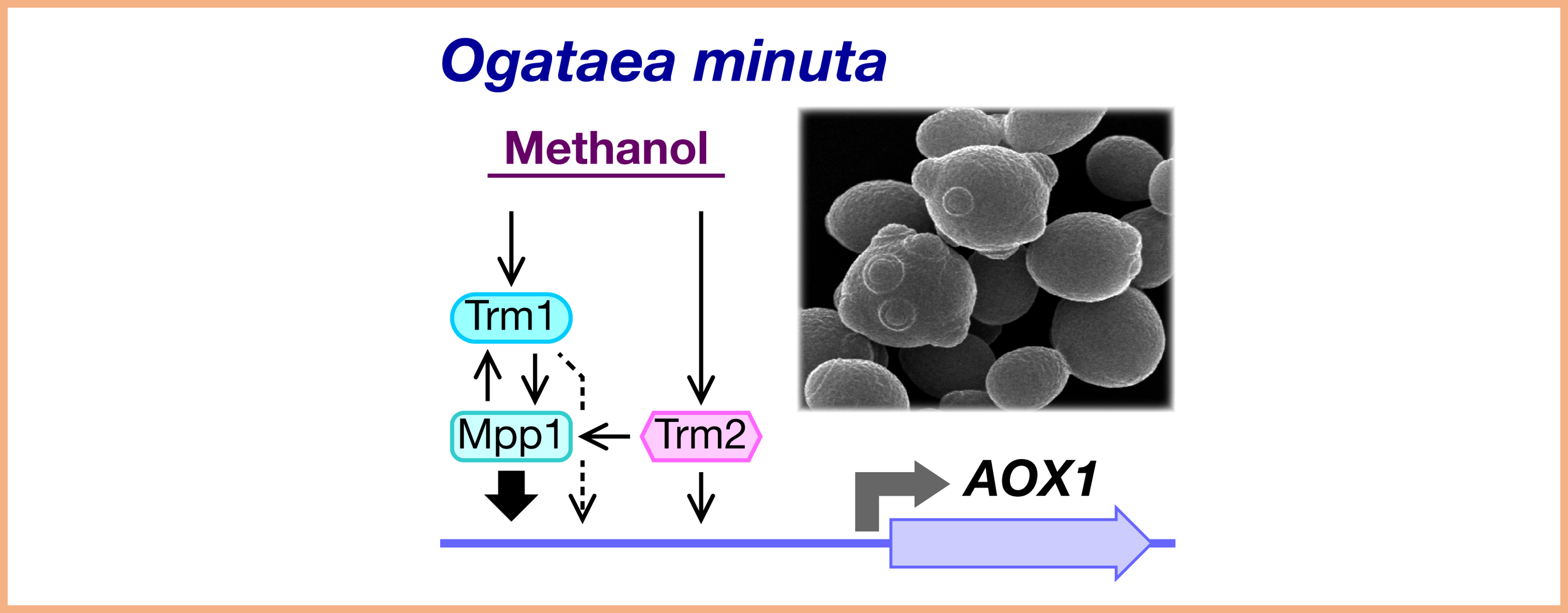
研究課題3:酵母による異種タンパク質生産
研究担当者:横尾 岳彦
メタノール資化性酵母 Ogataea minuta は、異種タンパク質生産に優れた宿主です。本種におけるプラスミド、ライブラリの開発や高分泌生産株創成に関する研究を行っています。また、グリコシルホスファチジルイノシトール(GPI)アンカーの合成機構や糖転移酵素の局在化機構に関する研究を行っています。
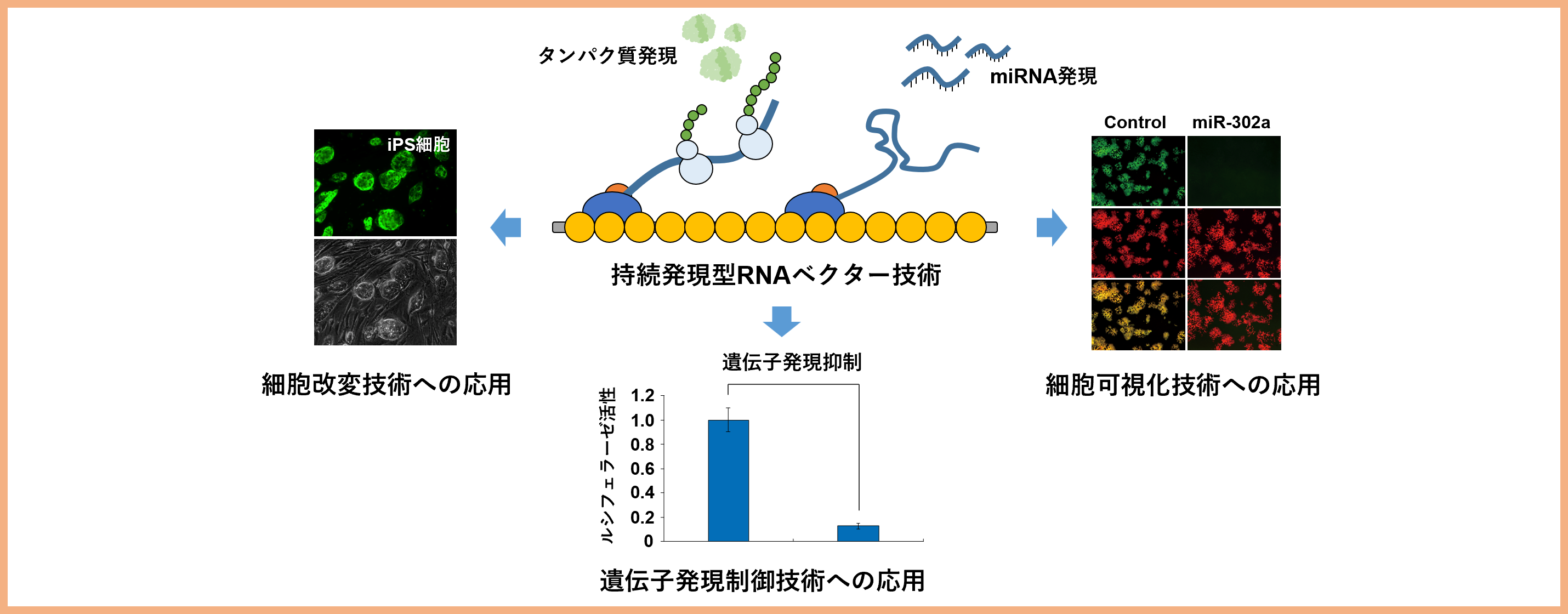
研究課題4:RNAベクターによる遺伝子導入技術の開発とその応用研究
研究担当者:佐野 将之
遺伝子導入技術には高い導入効率や発現持続性、導入した遺伝子の発現制御などが求められます。また、タンパク質だけでなくmiRNAのような非コードRNAを発現できることも必要になります。我々は、これらの特性を持つRNAベクターを開発することで、細胞の可視化や内在性遺伝子の発現制御、細胞の分化やリプログラミング制御など幅広い基礎・応用研究に利用しています。
グループの構成メンバー
| 顔写真 | 所属・役職および名前 | 専門分野 | その他、etc |
|---|---|---|---|
 |
グループ長 末永 光 |
|
|
 |
主任研究員 高橋 勝利 |
|
|
 |
主任研究員 佐野 将之 |
|
|
 |
上級主任研究員 今清水 正彦 |
|
|
 |
研究グループ付(兼務) 横尾 岳彦 |
|
業績リスト
- Zhu, J; Cai, HL; Goto, S; Shimada, M; Yurimoto, H; Sakai, Y; Yoko-O, T; Chiba, Y; Nakagawa, T.
Physiological role of the FDH1 gene in methylotrophic yeast Komagataella phaffii and carbon recovery as formate during methylotrophic growth of the fdh1-deletion strain.
J BIOSCI BIOENG. 2025 Jan 17. doi:10.1016/j.jbiosc.2024.12.007 - Hashimoto, T; Suenaga, H; Shin-Ya, K.
Application of Cas9-Based Gene Editing to Engineering of Nonribosomal Peptide Synthetases.
CHEMBIOCHEM. 2024 Dec 30:e202400765. doi:10.1002/cbic.202400765 - Kudo, K; Nishimura, T; Miyako, K; Suenaga, H; Shin-Ya, K.
A new cannabigerolic acid derivative and its unprenylated precursor produced through the reconstitution of cannabinoid biosynthesis in Streptomyces.
J ANTIBIOT (Tokyo). 2024 Dec 3. doi:10.1038/s41429-024-00793-5 - Yamamoto, K; Uzaki, M; Takahashi, K; Mimura, T.
Current status of MSI research in Japan to measure the localization of natural products in plants.
CURR OPIN PLANT BIOL. 2024 Oct 19;82:102651. doi: 10.1016/j.pbi.2024.102651 - Koshiguchi, M; Yonezawa, N; Hatano, Y; Suenaga, H; Yamagata, K; Kobayashi, S.
A system to analyze the initiation of random X-chromosome inactivation using time-lapse imaging of single cells.
SCI REP. 2024 Sep 2;14(1):20327. doi: 10.1038/s41598-024-71105-y - Song, X; Nihashi, Y; Imai, Y; Mori, N; Kagaya, N; Suenaga, H; Shin-Ya, K; Yamamoto, M; Setoyama, D; Kunisaki Y; Kida, YS .
Collagen Lattice Model, Populated with Heterogeneous Cancer-Associated Fibroblasts, Facilitates Advanced Reconstruction of Pancreatic Cancer Microenvironment.
INT J MOL SCI. 2024 Mar 27;25(7):3740. doi: 10.3390/ijms25073740 - Kudo, K; Nishimura, T; Izumikawa, M; Kozone, I; Hashimoto, J; Fujie, M; Suenaga, H; Ikeda, H; Satoh, N; Shin-Ya, K.
Capability of a large bacterial artificial chromosome clone harboring multiple biosynthetic gene clusters for the production of diverse compounds.
J ANTIBIOT (Tokyo). 2024 Mar 4. doi: 10.1038/s41429-024-00711-9 - Qayyum, MZ; Imashimizu, M; Leanca, M; Vishwakarma, RK; Riaz-Bradley, A; Yuzenkova, Y; Murakami, KS.
Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
PROC NATL ACAD SCI U S A. 2024 Feb 20;121(8):e2311480121. doi: 10.1073/pnas.2311480121 - Hashimoto, T; Suenaga, H; Amagai, K; Hashimoto, J; Kozone, I; Takahashi, S; Shin-Ya, K.
In Vitro Module Editing Of NRPS Enables Production Of Highly Potent Gq-Signaling Inhibitor FR900359 Derived From Unculturable Plant Symbiont.
ANGEW CHEM INT ED ENGL. 2024 Jan 18:e202317805. doi: 10.1002/anie.202317805 - Suzuki, S; Saito, S; Narushima, Y; Kodani, S; Kagaya, N; Suenaga, H; Shin-Ya, K; Arai, MA.
Notch activator cyclopiazonic acid induces apoptosis in HL-60 cells through calcineurin activation.
J ANTIBIOT (Tokyo). 2023 Nov 8. doi: 10.1038/s41429-023-00673-4 - Sugiyama, JI; Tokunaga, Y; Hishida, M; Tanaka, M; Takeuchi, K; Satoh, D; Imashimizu, M.
Nonthermal acceleration of protein hydration by sub-terahertz irradiation.
NAT COMMUN. 2023 May 22;14(1):2825. doi: 10.1038/s41467-023-38462-0 - Fujihara, H; Hirose, J; Suenaga, H.
Evolution of genetic architecture and gene regulation in biphenyl/PCB-degrading bacteria.
FRONTIERS IN MICROBIOLOGY. 2023 Jun 7;14:1168246. doi: 10.3389/fmicb.2023.1168246 - Tsuda, M; Nakatani, Y; Baba, S; Tanaka, I; Ichikawa, K; Nonaka, K; Ito, R; Yoko-O, T; Chiba, Y.
Selection of the methylotrophic yeast Ogataea minuta as a high-producing host for heterologous protein expression.
J BIOSCI BIOENG. 2023 Jan 24:S1389-1723(22)00375-9. doi: 10.1016/j.jbiosc.2022.12.006 - Choirunnisa, AR; Arima, K; Abe,Y; Kagaya, K; Kudo, K; Suenaga, H; Hashimoto, J; Fujie, M; Satoh, N; Shin-Ya, K; Matsuda, K; Wakimoto, T.
New azodyrecins identified by a genome mining-directed reactivity-based screening.
BEILSTEIN J ORG CHEM. 2022 Aug 10;18:1017-1025. doi: 10.3762/bjoc.18.102 - Mellul, M; Lahav, S; Imashimizu, M; Tokunaga, Y; Lukatsky, DB; Ram, O.
Repetitive DNA symmetry elements negatively regulate gene expression in embryonic stem cells.
BIOPHYS J. 2022 Jul 8:S0006-3495(22)00557-4. doi: 10.1016/j.bpj.2022.07.011 - Matsuda, K; Arima, K; Akiyama, S; Yamada, Y; Abe, Y; Suenaga, H; Hashimoto, J; Shin-Ya, K; Nishiyama, M; Wakimoto, T.
A Natural Dihydropyridazinone Scaffold Generated from a Unique Substrate for a Hydrazine-Forming Enzyme.
J AM CHEM SOC. 2022 Jun 30. doi: 10.1021/jacs.2c05269 - Yamamoto, K; Takahashi, K; OConnor, SE; Mimura, T.
Imaging MS Analysis in Catharanthus roseus.
METHODS MOL BIOL. 2022;2505:33-43. doi: 10.1007/978-1-0716-2349-7_2 - Suenaga, H; Matsuzawa, T; Sahara, T.
Discovery by metagenomics of a functional tandem repeat sequence that controls gene expression in bacteria.
FEMS MICROBIOL ECOL. 2022 Mar 28:fiac037. doi: 10.1093/femsec/fiac037 - Hirose, J; Watanabe, T; Futagami, T; Fujihara, H; Kimura, N; Suenaga, H; Goto, M; Suyama, A; Furukawa, K.
A New ICEclc Subfamily Integrative and Conjugative Element Responsible for Horizontal Transfer of Biphenyl and Salicylic Acid Catabolic Pathway in the PCB-Degrading Strain Pseudomonas stutzeri KF716.
MICROORGANISMS. 2021 Nov 29;9(12):2462. doi: 10.3390/microorganisms9122462 - Yoko-O, T; Komatsuzaki, A; Yoshihara, E; Zhao, S; Umemura, M; Gao, XD; Chiba, Y.
Regulation of alcohol oxidase gene expression in methylotrophic yeast Ogataea minuta.
J BIOSCI BIOENG. 2021 Aug 27:S1389-1723(21)00204-8. doi: 10.1016/j.jbiosc.2021.08.001 - Suenaga, H; Kagaya, N; Kawada, M; Tatsuda, D; Sato, T; Shin-Ya, K.
Phenotypic screening system using three-dimensional (3D) culture models for natural product screening.
J ANTIBIOT (Tokyo). 2021 Jul 29. doi: 10.1038/s41429-021-00457-8 - Nishimura, T; Kawahara, T; Kagaya, N; Ogura, Y; Takikawa, H; Suenaga, H; Adachi, M; Hirokawa, T; Doi, T; Shin-Ya, K.
JBIR-155, a Specific Class D β-Lactamase Inhibitor of Microbial Origin.
ORG LETT. 2021 May 24. doi: 10.1021/acs.orglett.1c01352 - Tokunaga, Y; Tanaka, M; Iida, H; Kinoshita, M; Tojima, Y; Takeuchi, K; Imashimizu, M.
Nonthermal excitation effects mediated by subterahertz radiation on hydrogen exchange in ubiquitin
BIOPHYS J. 2021 Apr 21:S0006-3495(21)00329-5. doi: 10.1016/j.bpj.2021.04.013