with Division Office
Research Project
Project 1:Development of Mechanical Analysis and Manipulation Techniques for Cells Using Nanomaterials
Researcher:NAKAMURA Chikashi
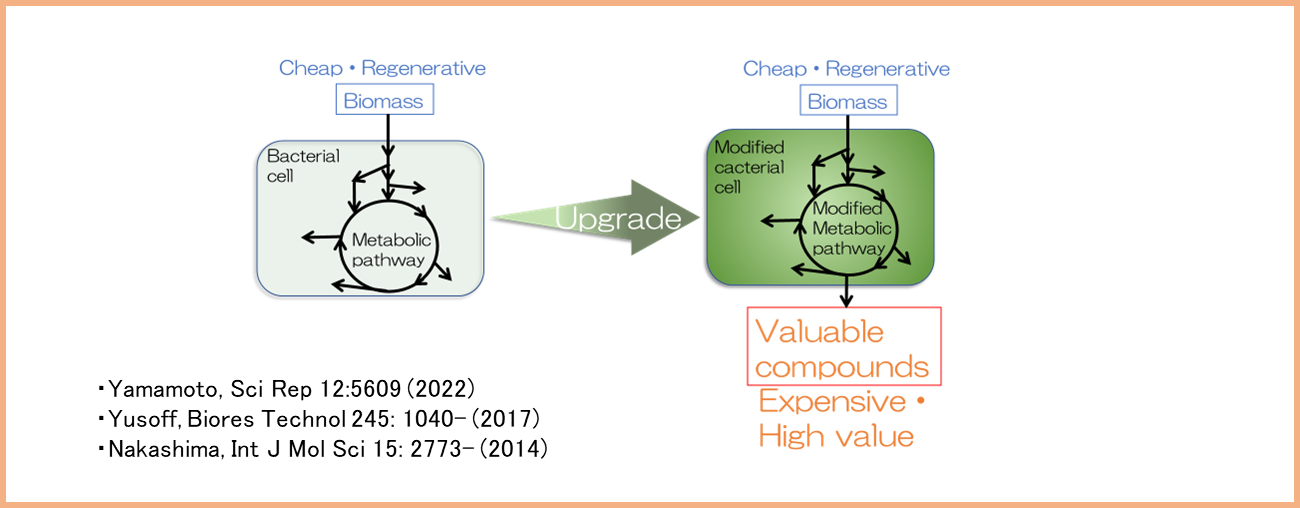
We are developing technologies that utilize non-damaging nanomaterials for the analysis and manipulation of living cells and organoids. This research is based on cell mechanics, which involves measuring forces from piconewtons to micronewtons using atomic force microscopy (AFM) to investigate interactions between antibodies and target membrane proteins, as well as the elasticity and adhesion forces of living cells. Specifically, our work includes genome editing using nanoneedles with a diameter of 200 nm, the construction of novel cancer models, and the development of therapeutic antibody drugs.
Project 2:Development of Advanced Human-Mimetic Evaluation Systems Using Microphysiological Systems, and Predictive Safety Assessment Models through the Digitalization of NAMs
Researcher:KIDA Yasuyuki
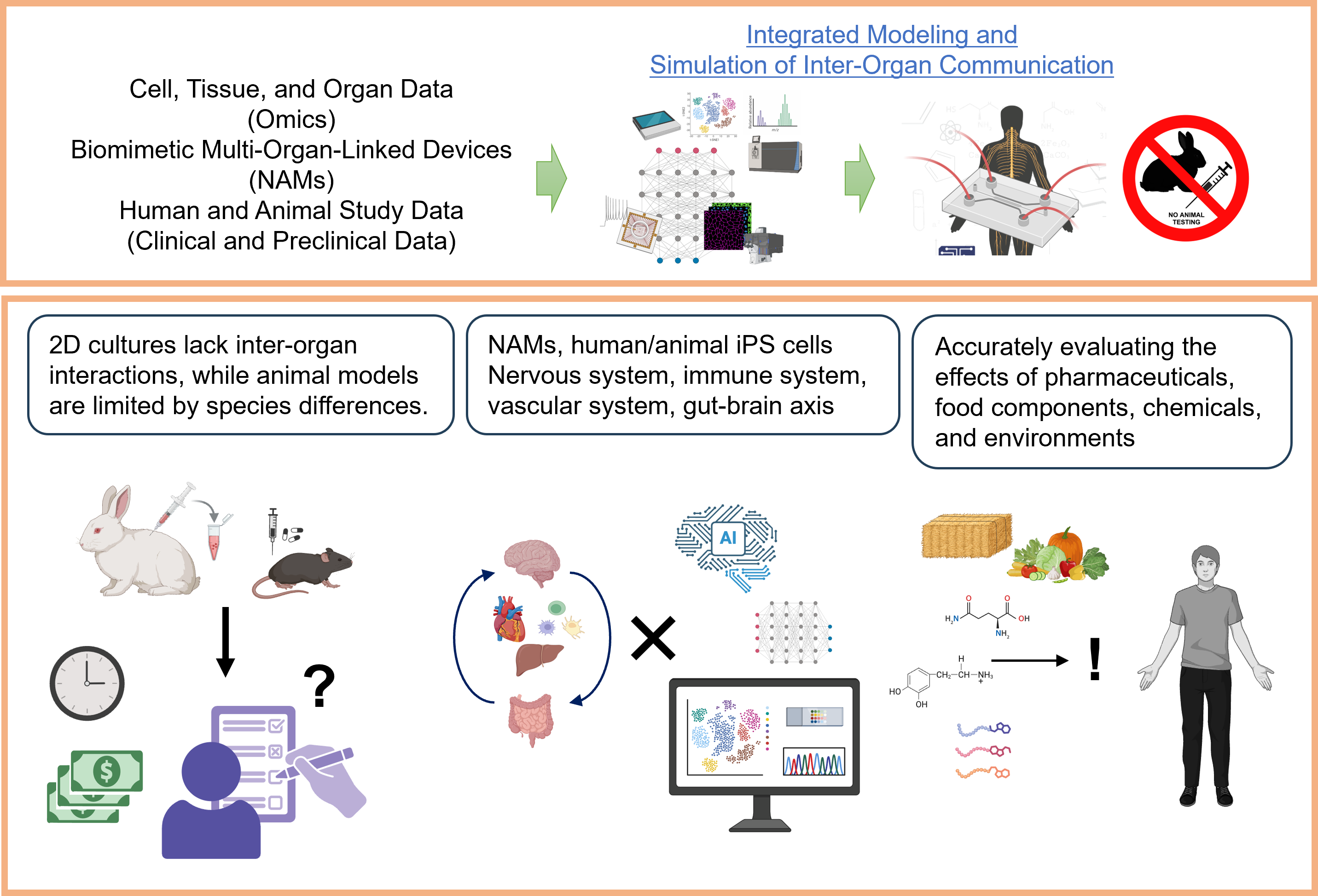
The advancement of bioassays for evaluating the safety and functionality of compounds is an urgent challenge. Traditionally, the evaluation of “biosystems”—referring to the mechanisms of biological responses, including physiological functions—has primarily relied on animal testing. However, a global shift toward alternative technologies is now underway, highlighting the need to establish a new academic research field tentatively referred to as biosystem replacement science. We are developing next-generation evaluation platforms with high human relevance, predictability, and reproducibility by integrating New Approach Methodologies (NAMs), omics analysis, and in silico simulations.
This research aims to build advanced evaluation systems that replicate the human physiological environment by integrating microphysiological systems (MPS) with diverse cells and tissues. These systems enable the analysis of physiological responses and drug mechanisms involving the autonomic nervous, immune, and vascular systems—areas difficult to assess with conventional models.
We combine MPS-based NAMs (New Approach Methodologies) with digital technologies such as omics data analysis and in silico simulations to develop predictive, data-driven safety assessment systems. Human iPS cells, along with those derived from animals like rats, mice, and rabbits, are used to create alternative evaluation platforms and cross-species comparative models. Our platform extends beyond pharmaceuticals and food, targeting chemical safety evaluation in housing and consumer products through in vitro × in silico approaches.
By enhancing prediction accuracy without relying on animal testing, we aim to support faster regulatory processes and more reliable product development. These technologies also contribute to the advancement and sustainability of bio-manufacturing and the wider bioeconomy.
Member
| photo | position & name | field of expertise | and other info |
|---|---|---|---|
 |
Principal research manager NAKAMURA Chikashi |
|
|
 |
Prime Senior Researcher NATSUME Toru |
|
|
 |
Attached to Research Institute KIDA Yasuyuki |
|
|
 |
Attached to Research Institute KIM Hyonchol |
|
|
 |
Career Researcher KAMEYAMA Akihiko |
|
|
Results
- Zeng, C; Hamada, M.
Identification and Sequence Characterization of Semi-extractable RNAs.
METHODS MOL BIOL. 2026;2949:239-252. doi:10.1007/978-1-0716-4670-0_15 - Tamai, K; Kinjo, S; Taguchi, A; Nagasaka, K; Yoshimoto, D; Duong, AQ; Yamamoto, Y; Iuchi, H; Mori, M; Sone, K; Hamada, M; Kawana, K; Ikeo, K; Hirota, Y; Osuga, Y.
Elucidating Alterations in Viral and Human Gene Expression Due to Human Papillomavirus Integration by Using Multimodal RNA Sequencing.
VIRUSES-BASEL. 2025 Oct 6;17(10):1344. doi:10.3390/v17101344 - Takeda, Y; Onoguchi, M; Ito, F; Yamamoto, I; Sumi, S; Yoshii, T; Ishida, M; Kajikawa, E; Zhang, J; Nishimura, O; Kadota, M; Tagami, S; Kondo, T; Saito, H; Hamada, M; Shibuya, H.
Nematode telomerase RNA hitchhikes on introns of germline-up-regulated genes.
SCIENCE. 2025 Oct 23;390(6771):eads7778. doi:10.1126/science.ads7778 - Hirano, Y; Kojima, N; Sakurai, H; Saito, E; Nankai, H; Komatsu, Y.
Analysis of the Effects of Sugar Modifications on RNA Chemical Ligation Reactions.
CHEMBIOCHEM. 2025 Oct 28:e2500263. doi:10.1002/cbic.202500263 - Ojima-Kato, T; Yokoyama, G; Nakano, H; Hamada, M; Motono, C.
Screening and machine learning-based prediction of translation-enhancing peptides that reduce ribosomal stalling in Escherichia coli.
RSC CHEM BIOL. 2025 Oct 22. doi:10.1039/d5cb00199d - Suzuki, T; Yasui, K; Takata, H; Komatsu, Y; Kamiya, H.
Action-at-a-Distance Mutations Induced by the True Abasic Site, Uracil, and Nicks: Unexpectedly Lower Mutagenicity of the True Abasic Site.
BIOLOGICAL & PHARMACEUTICAL BULLETIN. 2025;48(7):1111-1117. doi:10.1248/bpb.b25-00366 - Okazaki, T; Nozaki, K; Morimoto, N; Otobe, Y; Saito, R; Abe, S; Okajima, M; Yoshitane, H; Hatta, T; Iemura, SI; Natsume, T; Kosako, H; Yamasaki, M; Inoue, S; Kondo, T; Koseki, H; Gotoh, Y.
Membrane topology inversion of GGCX mediates cytoplasmic carboxylation for antiviral defense.
SCIENCE. 2025 Jul 3;389(6755):84-91. doi: 10.1126/science.adk9967 - Soeda, S; Montrose, M; Takahashi, A; Ishida, R; Burriel, S; Ohmine, N; Natsume, T; Adachi, S; Kim, M; Yamamoto, T.
The E3 ubiquitin ligase, RNF219, suppresses CNOT6L expression to exhibit antiproliferative activity.
FEBS OPEN BIO. 2025 Jul 1. doi: 10.1002/2211-5463.70081 - Nakano, R; Iwano, N; Ichinose, A; Hamada, M.
RaptGen-UI: an integrated platform for exploring and analyzing the sequence landscape of HT-SELEX experiments.
BIOINFORM ADV. 2025 May 23;5(1):vbaf120. doi: 10.1093/bioadv/vbaf120 - Kawai, Y; Kameyama, A; Sakuraba, H; Matsuzawa, T.
Characterization of glycoside hydrolases involved in xyloglucan degradation in the thermophilic bacterium Thermotoga maritima.
SCIENTIFIC REPORTS. 2025 Jun 6;15(1):19922. doi:10.1038/s41598-025-05366-6 - Date, Y; Sasazawa, Y; Kitagawa, M; Gejima, K; Suzuki, A; Saya, H; Kida, Y; Imoto, M; Itakura, E; Hattori, N; Saiki, S.
Novel autophagy inducers by accelerating lysosomal clustering against Parkinsons disease.
ELIFE. 2024 Jul 3:13:e98649. doi:10.7554/eLife.98649 - Mori, N; Miyazaki, Y; Oda, T; Kida, YS.
RT-4M: Real-Time Mosaicing Manager for Manual Microscopy System.
Sensors (Basel). 2025 May 8;25(10):2968. doi:10.3390/s25102968 - Yamada, K; Suga, K; Abe, N; Hashimoto, K; Tsutsumi, S; Inagaki, M; Hashiya, F; Abe, H; Hamada, M.
Multi-objective computational optimization of human 5 UTR sequences.
BRIEF BIOINFORM. 2025 May 1;26(3):bbaf225. doi:10.1093/bib/bbaf225 - Nobe, M; Maruzuru, Y; Takeshima, K; Maeda, F; Kusano, H; Yoshimura, R; Nishiyama, T; Park, H; Kozaki, Y; Iwami, S; Koyanagi, N; Kato, A; Natsume, T; Adachi, S; Kawaguchi, Y.
Direct relationship between protein expression and progeny yield of herpes simplex virus 1.
MBIO. 2025 May 5:e0028025. doi:10.1128/mbio.00280-25 - Takeda, A; Nonaka, D; Imazu, Y; Fukunaga, T; Hamada, M.
REPrise: de novo interspersed repeat detection using inexact seeding.
MOB DNA. 2025 Apr 3;16(1):16. doi:10.1186/s13100-025-00353-0 - Ishii, N; Koja, Y; Noguchi, K; Yohda, M; Takeda, S.
Enhancing specimen preparation for transmission electron microscopy: Trypan Blue staining and low-melting-point agar embedding for ultra-thin cell sections.
J BIOSCI BIOENG. 2025 Mar 6:S1389-1723(25)00029-5. doi:10.1016/j.jbiosc.2025.02.003 - Ishii, N; Tateno, H.
Preparation of Small Extracellular Vesicles Using Sequential Ultrafiltration with Regenerated Cellulose Membranes of Different Molecular Weight Cutoffs: A Study of Morphology and Size by Electron Microscopy.
MICROSC MICROANAL. 2025 Jan 13. doi:10.1093/mam/ozae133 - Ishii, N; Ogawa, Y.
Grids designed for tomography: Stereovision transmission electron microscopy makes it easy to determine the winding handedness of helical nanocoils.
MICRON. 2025 Mar;190:103784. Epub 2025 Jan 7. doi:10.1016/j.micron.2025.103784 - Hossain, AS; Clarin, MTRDC; Kimura, K; Biggin, G; Taga, Y; Uto, K; Yamagishi, A; Motoyama, E; Narenmandula; Mizuno, K; Nakamura, C; Asano, K; Ohtsuki, S; Nakamura, T; Kanki, S; Baldock, C; Raja, E; Yanagisawa, H.
Fibrillin-1 G234D mutation in the hybrid1 domain causes tight skin associated with dysregulated elastogenesis and increased collagen cross-linking in mice.
MATRIX BIOL. 2024 Nov 28:S0945-053X(24)00142-2. doi:10.1016/j.matbio.2024.11.006 - Kakizuka, T; Natsume, T; Nagai, T.
Compact lens-free imager using a thin-film transistor for long-term quantitative monitoring of stem cell culture and cardiomyocyte production.
LAB ON A CHIP. 2024 OCT 14. doi: 10.1039/d4lc00528g - Morikawa, K; Nagasaki, A; Sun, L; Kawase, E; Ebihara, T; Shirayoshi, Y.
Optogenetic control of early embryos labeling using photoactivatable Cre recombinase 3.0.
FEBS OPEN BIO. 2024 Sep 2. doi: 10.1002/2211-5463.13862 - Tokuoka, SM; Hamano, F; Kobayashi, A; Adachi, S; Andou, T; Natsume, T; Oda, Y.
Plasma proteomics and lipidomics facilitate elucidation of the link between Alzheimers disease development and vessel wall fragility.
SCI REP. 2024 Aug 27;14(1):19901. doi: 10.1038/s41598-024-71097-9 - Akagi, Y; Norimoto, A; Kawamura, T; Kida, YS.
Label-Free Assessment of Neuronal Activity Using Raman Micro-Spectroscopy.
MOLECULES. 2024 Jul 3;29(13):3174. doi: 10.3390/molecules29133174 - Shimada, N; Kameyama, A; Watanabe, M; Sahara, T; Matsuzawa, T.
Identification and characterization of xyloglucan-degradation related α-1,2-l-fucosidase in Aspergillus oryzae.
J BIOSCI BIOENG. 2024 Jun 12:S1389-1723(24)00159-2. doi: 10.1016/j.jbiosc.2024.05.011 - Kato, A; Iwasaki, R; Takeshima, K; Maruzuru, Y; Koyanagi, N; Natsume, T; Kusano, H; Adachi, S; Kawano, S; Kawaguchi, Y.
Identification of a novel neurovirulence factor encoded by the cryptic orphan gene UL31.6 of herpes simplex virus 1.
J VIROL. 2024 May 31:e0074724. doi: 10.1128/jvi.00747-24 - Song, X; Nihashi, Y; Imai, Y; Mori, N; Kagaya, N; Suenaga, H; Shin-Ya, K; Yamamoto, M; Setoyama, D; Kunisaki Y; Kida, YS .
Collagen Lattice Model, Populated with Heterogeneous Cancer-Associated Fibroblasts, Facilitates Advanced Reconstruction of Pancreatic Cancer Microenvironment.
INT J MOL SCI. 2024 Mar 27;25(7):3740. doi: 10.3390/ijms25073740 - Kanie, S; Wu, C; Kihira, K; Yasuno, R; Mitani, Y; Ohmiya, Y.
Bioluminescence of (R)-Cypridina Luciferin with Cypridina Luciferase.
INT J MOL SCI. 2024 Feb 26;25(5):2699. doi: 10.3390/ijms25052699 - Sakata, S; Li, J; Yasuno, Y; Shinada, T; Shin-ya, K;Katsuyama, Y; Ohnishi, Y.
Identification of the cirratiomycin biosynthesis gene cluster in Streptomyces cirratus: elucidation of the biosynthetic pathways for 2,3-diaminobutyric acid and hydroxymethylserine.
CHEMISTRY. 2024 Mar 8:e202400271. doi: 10.1002/chem.202400271 - Zheng, Y; Morita, N; Takagi, H; Shiozaki-Sato, Y; Ishikawa, J; Shin-ya, K;Takahashi, S.
Alanyl-tRNA Synthetase-like Enzyme-Catalyzed Aminoacylation in Nucleoside Sulfamate Ascamycin Biosynthesis.
ACS CATALYSIS. 2024 FEB 20. doi: 10.1021/acscatal.3c05667 - Kudo, K; Nishimura, T; Izumikawa, M; Kozone, I; Hashimoto, J; Fujie, M; Suenaga, H; Ikeda, H; Satoh, N; Shin-Ya, K.
Capability of a large bacterial artificial chromosome clone harboring multiple biosynthetic gene clusters for the production of diverse compounds.
J ANTIBIOT (Tokyo). 2024 Mar 4. doi: 10.1038/s41429-024-00711-9 - Sun, L; Morikawa, K; Sogo, Y; Sugiura, Y.
MHY1485 potentiates immunogenic cell death induction and anti-cancer immunity following irradiation.
J RADIAT RES. 2024 Feb 8:rrad107. doi: 10.1093/jrr/rrad107 - Dong, W; Kameyama, A.
Succinylation-Alcian Blue Staining of Mucins on Polyvinylidene Difluoride Membrane.
METHODS MOL BIOL. 2024;2763:111-117. doi: 10.1007/978-1-0716-3670-1_9 - Kameyama, A.
Eliminative Oximation of O-Glycans from Mucins.
METHODS MOL BIOL. 2024;2763:151-158. doi: 10.1007/978-1-0716-3670-1_13 - Kameyama, A.
Supported Molecular Matrix Electrophoresis.
METHODS MOL BIOL. 2024;2763:79-97. doi: 10.1007/978-1-0716-3670-1_7 - Sugiura, T; Kameyama, A.
Preparation of Soluble Mucin Solutions from the Salivary Glands.
METHODS MOL BIOL. 2024;2763:45-50. doi: 10.1007/978-1-0716-3670-1_3 - Hashimoto, T; Suenaga, H; Amagai, K; Hashimoto, J; Kozone, I; Takahashi, S; Shin-Ya, K.
In Vitro Module Editing Of NRPS Enables Production Of Highly Potent Gq-Signaling Inhibitor FR900359 Derived From Unculturable Plant Symbiont.
ANGEW CHEM INT ED ENGL. 2024 Jan 18:e202317805. doi: 10.1002/anie.202317805 - Song, X; Nihashi, Y; amamoto, M; Setoyama, D; Kunisaki, Y; Kida, YS.
Exploring the Role of Desmoplastic Physical Stroma in Pancreatic Cancer Progression Using a Three-Dimensional Collagen Matrix Model.
BIOENGINEERING (Basel). 2023 Dec 18;10(12):1437. doi: 10.3390/bioengineering10121437 - Akagi, Y; Takayama, Y; Nihashi, Y; Yamashita, A; Yoshida, R; Miyamoto, Y; Kida, YS.
Functional engineering of human iPSC-derived parasympathetic neurons enhances responsiveness to gastrointestinal hormones.
FEBS OPEN BIO. 2023 Nov 27. doi: 10.1002/2211-5463.13741 - Suzuki, S; Saito, S; Narushima, Y; Kodani, S; Kagaya, N; Suenaga, H; Shin-Ya, K; Arai, MA.
Notch activator cyclopiazonic acid induces apoptosis in HL-60 cells through calcineurin activation.
J ANTIBIOT (Tokyo). 2023 Nov 8. doi: 10.1038/s41429-023-00673-4 - Takakuwa, H; Yamazaki, T; Souquere, S; Adachi, S; Yoshino, H; Fujiwara, N; Yamamoto, T; Natsume, T; Nakagawa, S; Pierron, G; Hirose, T.
Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles.
NAT CELL BIOL. 2023 Nov 6. doi: 10.1038/s41556-023-01254-1 - Ishii, N; Noguchi, K; Ikemoto, MJ; Yohda, M; Odahara, T.
Optimizing Exosome Preparation Based on Size and Morphology: Insights From Electron Microscopy.
MICROSC MICROANAL. 2023 Oct 13:ozad103. doi: 10.1093/micmic/ozad103 - Ishii, N; Odahara, T.
Investigation of the Efficacy of Lanthanoid Heavy Metal Acetates as Electron Staining Reagents for Biomembrane Vesicles.
MICROSC MICROANAL. 2023 Oct 13:ozad107. doi: 10.1093/micmic/ozad107 - Ishioka, M; Nihashi, Y; Sunagawa, Y; Umezawa, K; Shimosato, T; Kagami, H; Morimoto, T; Takaya, T.
Myogenetic Oligodeoxynucleotide Induces Myocardial Differentiation of Murine Pluripotent Stem Cells.
INT J MOL SCI. 2023 Sep 21;24(18):14380. doi: 10.3390/ijms241814380 - Miyazawa, K; Penedo, M; Furusho, H; Ichikawa, T; Alam, MS; Miyata, K; Nakamura, C; Fukuma, T.
Nanoendoscopy-AFM for Visualizing Intracellular Nanostructures of Living Cells.
MICROSC MICROANAL. 2023 Jul 22;29(Supplement_1):782. doi: 10.1093/micmic/ozad067.387 - Takubo, K; Htun, PW; Ueda, T; Sera, Y; Iwasaki, M; Koizumi, M; Shiroshita, K; Kobayashi, H; Haraguchi, M; Watanuki, S; Honda, ZI; Yamasaki, N; Nakamura-Ishizu, A; Arai, F; Motoyama, N; Hatta, T; Natsume, T; Suda, T; Honda, H.
MBTD1 preserves adult hematopoietic stem cell pool size and function.
PROC NATL ACAD SCI U S A. 2023 Aug 8;120(32):e2206860120. doi: 10.1073/pnas.2206860120 - Ichikawa, T; Alam, MS; Penedo, M; Matsumoto, K; Fujita, S; Miyazawa, K; Furusho, H; Miyata, K; Nakamura, C; Fukuma, T.
Protocol for live imaging of intracellular nanoscale structures using atomic force microscopy with nanoneedle probes.
STAR PROTOC. 2023 Jul 22;4(3):102468. doi: 10.1016/j.xpro.2023.102468 - Miyashita, K; Yagi, T; Kagaya, N; Takechi, A; Nakata, C; Kanda, R; Nuriya, H; Tanegashima, K; Hoyano, S; Seki, F; Yoshida, C; Hachiro, Y; Higashi, T; Kitada, N; Toya, T; Kobayashi, T; Najima, Y; Goyama, S; Maki, SA; Kitamura, T; Doki, N; Shin-Ya, K; Hara, T.
Identification of compounds that preferentially suppress the growth of T-cell acute lymphoblastic leukemia-derived cells.
CANCER SCI. 2023 Jul 31. doi: 10.1111/cas.15918 - Nihashi, Y, Song, X; Yamamoto, M; Setoyama, D; Kida, YS.
Decoding Metabolic Symbiosis between Pancreatic Cancer Cells and Cancer-Associated Fibroblasts Using Cultured Tumor Microenvironment.
INT J MOL SCI. 2023 Jul 3;24(13):11015. doi: 10.3390/ijms241311015 - Taniguchi, M; Okumura, R; Matsuzaki, T; Nakatani, A; Sakaki, K; Okamoto, S; Ishibashi, A; Tani, H; Horikiri, M; Kobayashi, N; Yoshikawa, HY; Motooka, D; Okuzaki, D; Nakamura, S; Kida, T; Kameyama, A, Takeda, K.
Sialylation shapes mucus architecture inhibiting bacterial invasion in the colon.
MUCOSAL IMMUNOL. 2023 Jun 27:S1933-0219(23)00049-1. doi: 10.1016/j.mucimm.2023.06.004 - Roberts, MD; McCarthy, JJ; Hornberger, TA; Phillips, SM; Mackey, AL; Nader, GA; Boppart, MD; Kavazis, AN; Reidy, PT; Ogasawara, R, Libardi, CA; Ugrinowitsch, C; Booth, FW; Esser, KA.
Mechanisms of mechanical overload-induced skeletal muscle hypertrophy: current understanding and future directions.
PHYSIOL REV. 2023 Jun 29. doi: 10.1152/physrev.00039.2022 - Zhuang, HT; Fujikura, Y, Ohkura, N; Higo-Yamamoto, S; Mishima, T; Oishi, K.
A ketogenic diet containing medium-chain triglycerides reduces REM sleep duration without significant influence on mouse circadian phenotypes.
FOOD RESEARCH INTERNATIONAL. 2023 Jul;169:112852. doi: 10.1016/j.foodres.2023.112852 - Takayama, Y, Akagi, Y, Kida, YS.
Deciphering the Molecular Mechanisms of Autonomic Nervous System Neuron Induction through Integrative Bioinformatics Analysis.
INT J MOL SCI. 2023 May 21;24(10):9053. doi: 10.3390/ijms24109053 - Arima, K; Akiyama, S; Shin-Ya, K; Matsuda, K; Wakimoto, T.
Carrier Protein Mediated Formation of the Dihydropyridazinone Ring in Actinopyridazinone Biosynthesis.
ANGEW CHEM INT ED ENGL. 2023 May 17:e202305155. doi: 10.1002/anie.202305155 - Kumagai, Y; Saito, Y; Kida, YS.
A multiomics atlas of brown adipose tissue development over time.
ENDOCRINOLOGY. 2023 Apr 21:bqad064. doi: 10.1210/endocr/bqad064 - Sugiura, T; Hashimoto, K; Kikuta, K; Anazawa, U; Nomura, T; Kameyama, A.
Expression and localisation of MUC1 modified with sialylated core-2 O-glycans in mucoepidermoid carcinoma.
SCI REP. 2023 Apr 8;13(1):5752. doi: 10.1038/s41598-023-32597-2 - Ikemoto, MJ; Aihara, Y; Ishii, N; Shigemori, H.
3,4-Dihydroxybenzalacetone Inhibits the Propagation of Hydrogen Peroxide-Induced Oxidative Effect via Secretory Components from SH-SY5Y Cells.
BIOL PHARM BULL. 2023;46(4):599-607. doi: 10.1248/bpb.b22-00727 - Murata, Y; Jitsukawa, T; Iida, M; Furuta, T; Haramoto, Y; Shigeri, Y; Fujisaki, S.
Liquid chromatography-mass spectrometry analyses of polyprenyl phosphates in Escherichia coli cells in which genes for isoprenoid synthesis are amplified.
J BIOSCI BIOENG. 2023 Mar 1:S1389-1723(23)00047-6. doi: 10.1016/j.jbiosc.2023.02.003 - Sogo, T; Nakao, S; Tsukamoto, T; Ueyama, T; Harada, Y; Ihara, D; Ishida, T; Nakahara, M; Hasegawa, K; Akagi, Y; Kida, YS; Nakagawa, O; Nagamune, T; Kawahara, M; Kawamura, T.
Canonical Wnt signaling activation by chimeric antigen receptors for efficient cardiac differentiation from mouse embryonic stem cells.
INFLAMM REGEN. 2023 Feb 10;43(1):11. doi: 10.1186/s41232-023-00258-6 - Ato, S; Matsunami, H;Ogasawara, R.
Aging is associated with impaired postprandial response of skeletal muscle protein synthesis to high-intensity muscle contraction in mice.
J GERONTOL A BIOL SCI MED SCI. 2023 Jan 12:glad014. doi: 10.1093/gerona/glad014 - Mawaribuchi, S; Haramoto, Y; Ikeda, N; Ito, M.
Evolutionary features of ligands and their receptors via protein-protein interactions and essentiality in primates.
GENES CELLS. 2023 Jan 9. doi: 10.1111/gtc.13006 - Kudo, F; Kishikawa, K; Tsuboi, K; Kido, T; Usui, T; Hashimoto, J; Shin-Ya, K; Miyanaga, A; Eguchi, T.
Acyltransferase Domain Exchange between Two Independent Type I Polyketide Synthases in the Same Producer Strain of Macrolide Antibiotics.
CHEMBIOCHEM. 2023 Jan 5. doi: 10.1002/cbic.202200670 - Ishii, N.
C-shaped dipper: A novel useful auxiliary tool for preparation of specimen grids for transmission electron microscopy.
MICROSC RES TECH. 2022 Dec 29. doi: 10.1002/jemt.24283 - Ishimura, R; El-Gowily, AH; Noshiro, D; Komatsu-Hirota, S; Ono, Y; Shindo, M; Hatta, T; Abe, M; Uemura, T; Lee-Okada, HC; TMohamed, TM; Yokomizo, T; Ueno, T; Sakimura, K; Natsume, T; Sorimachi, H; Inada, T; Waguri, S; Noda, NN; Komatsu, M.
The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
NAT COMMUN. 2022 Dec 21;13(1):7857. doi: 10.1038/s41467-022-35501-0 - Ando, M; Kawasaki, H; Tamura, S; Haramoto, Y; Shigeri, Y.
Recent Advances in Gas Sensing Technology Using Non-Oxide II-VI Semiconductors CdS, CdSe, and CdTe.
CHEMOSENSORS. 2022 10(11)482. doi: 10.3390/chemosensors10110482 - Yoshida, Y; Yajima, Y; Fujikura, Y; Zhuang, H; Higo-Yamamoto, S; Toyoda, A; Oishi, K.
Identification of salivary microRNA profiles in male mouse model of chronic sleep disorder.
Stress. 2022;26(1):21-28. doi: 10.1080/10253890.2022.2156783 - Haji, S; Ito, T; Guenther, C; Nakano, M; Shimizu, T; Mori, D; Chiba, Y; Tanaka, M; Mishra, SK; Willment, JA; Brown, GD; Nagae, M; Yamasaki, S.
Human Dectin-1 is O-glycosylated and serves as a ligand for C-type lectin receptor CLEC-2.
ELIFE. 2022 Dec 8;11:e83037. doi: 10.7554/eLife.83037 - Kudo, F; Minato, A; Sato, S; Nagano, N; Maruyama, C; Hamano, Y; Hashimoto, J; Kozone, I; Shin-Ya; Eguchi, T.
Mechanism of S-Adenosyl-l-methionine C-Methylation by Cobalamin-dependent Radical S-Adenosyl-l-methionine Methylase in 1-Amino-2-methylcyclopropanecarboxylic Acid Biosynthesis.
ORG LETT. 2022 Dec 2. doi: 10.1021/acs.orglett.2c03555 - Kato, K; Okazaki, S; Schmitt-Ulms, C; Jiang, K; Zhou, W; Ishikawa, J; Isayama, Y; Adachi, S; Nishizawa, T; Makarova, KS; Koonin, EV; Abudayyeh, OO; Gootenberg, JS; Nishimasu, H.
RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
SCIENCE. 2022 Nov 25;378(6622):882-889. doi: 10.1126/science.add7347 - Fujikura, Y; Yamanouchi, K; Sugihara, H; Hatakeyama, M; Abe, T; Ato, S; Oishi, K.
Ketogenic diet containing medium-chain triglyceride ameliorates transcriptome disruption in skeletal muscles of rat models of duchenne muscular dystrophy.
BIOCHEM BIOPHYS REP. 2022 Nov 11;32:101378. doi: 10.1016/j.bbrep.2022.101378 - Nohira, N; Shinji, S; Nakamura, S; Nihashi, Y; Shimosato, T; Takaya, T.
Myogenetic Oligodeoxynucleotides as Anti-Nucleolin Aptamers Inhibit the Growth of Embryonal Rhabdomyosarcoma Cells.
BIOMEDICINES. 2022 Oct 25;10(11):2691. doi: 10.3390/biomedicines10112691 - Imai, Y;Mori, N; Nihashi, Y; Kumagai, Y; Shibuya, Y; Oshima, J; Sasaki, M; Sasaki, K; Aihara, Y; Sekido, M; Kida, YS.
Therapeutic Potential of Adipose Stem Cell-Derived Conditioned Medium on Scar Contraction Model.
BIOMEDICINES. 2022 Sep 24;10(10):2388. doi: 10.3390/biomedicines10102388 - Tirta, YK; Adachi, S; Perez, CAG; Adhitama, N; Nong, QD; Natsume, T; Kato, Y; Watanabe, H.
CELF1 represses Doublesex1 expression via its 5 UTR in the crustacean Daphnia magna.
PLOS ONE. 2022 Oct 14;17(10):e0275526. doi: 10.1371/journal.pone.0275526 - Sato, R; Suzuki, K; Yasuda, Y; Suenaga, A; Fukui, K.
RNAapt3D: RNA aptamer 3D-structural modeling database.
BIOPHYS J. 2022 Sep 22:S0006-3495(22)00773-1. doi: 10.1016/j.bpj.2022.09.023 - Kawai, S; Hagihara, R; Shin-Ya, K; Katsuyama, Y; Ohnishi, Y.
Bacterial Avenalumic Acid Biosynthesis Includes Substitution of an Aromatic Amino Group for Hydride by Nitrous Acid Dependent Diazotization.
ANGEW CHEM INT ED ENGL. 2022 Sep 17. doi: 10.1002/anie.202211728 - Teranishi, H; Tabata, K; Saeki, M; Umemoto, T; Hatta, T; Otomo, T; Yamamoto, K; Natsume, T; Yoshimori, T; Hamasaki, M.
Identification of CUL4A-DDB1-WDFY1 as an E3 ubiquitin ligase complex involved in initiation of lysophagy.
CELL REP. 2022 Sep 13;40(11):111349. doi: 10.1016/j.celrep.2022.111349 - Li, S; Ikeuchi, K; Kato, M; Buschauer, R; Sugiyama, T; Adachi, S; Kusano, H; Natsume, T; Berninghausen, O; Matsuo, Y; Becker, T; Beckmann, R; Inada, T.
Sensing of individual stalled 80S ribosomes by Fap1 for nonfunctional rRNA turnover.
MOL CELL. 2022 Sep 15;82(18):3424-3437.e8. doi: 10.1016/j.molcel.2022.08.018 - Kagiwada, H; Motono, C; Horimoto, K; Fukui, K.
Phosprof: pathway analysis database of drug response based on phosphorylation activity measurements.
DATABASE (Oxford). 2022 Aug 22;2022:baac072. doi: 10.1093/database/baac072 - Yamagishi, A; Mizusawa, M; Uchida, K; Iijima, M; Kuroda, S; Fukazawa, K; Ishihara, K; Nakamura, C.
Mechanical detection of interactions between proteins related to intermediate filament and transcriptional regulation in living cells.
BIOSENS BIOELECTRON. 2022 Aug 6;216:114603. doi: 10.1016/j.bios.2022.114603 - Kokubu, R; Ohno, S; Kuratani, H; Takahashi, Y; Manabe, N; Shimizu, H; Chiba, Y; Denda-Nagai, K; Tsuiji, M; Irimura, T; Yamaguchi, Y.
O-Glycan-Dependent Interaction between MUC1 Glycopeptide and MY.1E12 Antibody by NMR, Molecular Dynamics and Docking Simulations.
INT J MOL SCI. 2022 Jul 16;23(14):7855. doi: 10.3390/ijms23147855 - Iwamura, C; Hirahara, K; Kiuchi, M; Ikehara, S; Azuma, K; Shimada, T; Kuriyama, S; Ohki, S; Yamamoto, E; Inaba, Y; Shiko, Y; Aoki, A; Kokubo, K; Hirasawa, R; Hishiya, T; Tsuji, K; Nagaoka, T; Ishikawa, S; Kojima, A; Mito, H; Hase, R; Kasahara, Y; Kuriyama, N; Tsukamoto, T; Nakamura, S; Urushibara, T; Kaneda, S; Sakao, S; Tobiume, M; Suzuki, Y; Tsujiwaki, M; Kubo, T; Hasegawa, T; Nakase, H; Nishida, O; Takahashi, K; Baba, K; Iizumi, Y; Okazaki, T; Kimura, MY; Yoshino, I; Igari, H; Nakajima, H; Suzuki, T; Hanaoka, H; Nakada, TA; Ikehara, Y; Yokote, K; Nakayama, T.
Elevated Myl9 reflects the Myl9-containing microthrombi in SARS-CoV-2-induced lung exudative vasculitis and predicts COVID-19 severity.
PROC NATL ACAD SCI USA. 2022 Aug 16;119(33):e2203437119. doi: 10.1073/pnas.2203437119 - Yasuno, R; Mitani, Y; Ohmiya, Y.
Gene Cloning and Functional Analysis of the Luciferase from Luminous Syllids of the Genus Odontosyllis.
METHODS MOL BIOL. 2022;2524:3-15. doi: 10.1007/978-1-0716-2453-1_1 - Fujikura, Y; Kimura, K; Yamanouchi, K; Sugihara, H; Hatakeyama, M; Zhuang, H; Abe, T; Daimon, M; Morita, H; Komuro, I; Oishi, K.
A medium-chain triglyceride containing ketogenic diet exacerbates cardiomyopathy in a CRISPR/Cas9 gene-edited rat model with Duchenne muscular dystrophy.
SCI REP. 2022 Jul 8;12(1):11580. doi: 10.1038/s41598-022-15934-9 - Viswan, A; Yamagishi, A; Hoshi, M; Furuhata, Y; Kato, Y; Makimoto, N; Takeshita, T; Kobayashi, T; Iwata, F; Kimura, M; Yoshizumi, T; Nakamura, C.
Microneedle Array-Assisted, Direct Delivery of Genome-Editing Proteins Into Plant Tissue.
FRONT PLANT SCI. 2022 Jun 24;13:878059. doi: 10.3389/fpls.2022.878059 - Matsuda, K; Arima, K; Akiyama, S; Yamada, Y; Abe, Y; Suenaga, H; Hashimoto, J; Shin-Ya, K; Nishiyama, M; Wakimoto, T.
A Natural Dihydropyridazinone Scaffold Generated from a Unique Substrate for a Hydrazine-Forming Enzyme.
J AM CHEM SOC. 2022 Jun 30. doi: 10.1021/jacs.2c05269 - Kanda, GN; Tsuzuki, T; Terada, M; Sakai, N; Motozawa, N; Masuda, T; Nishida, M; Watanabe, CT; Higashi, T; Horiguchi, SA; Taku Kudo, T; Kamei, M; Sunagawa, GA; Matsukuma, K; Sakurada, T; Ozawa, Y; Takahashi, M; Takahashi, K; Natsume, T.
Robotic search for optimal cell culture in regenerative medicine.
ELIFE. 2022 Jun 28;11:e77007. doi: 10.7554/eLife.77007 - Sasaki, K; Suzuki, M; Sonoda, T; Schneider-Poetsch, T; Ito, A; Takagi, M; Fujishiro, S; Sohtome, Y; Dodo, K; Umehara, T; Aburatani, H; Shin-Ya, K; Nakao, Y; Sodeoka, M; Yoshida, M.
Visualization of the dynamic interaction between nucleosomal histone H3K9 tri-methylation and HP1α chromodomain in living cells.
CELL CHEM BIOL. 2022 Jun 18:S2451-9456(22)00198-2. doi: 10.1016/j.chembiol.2022.05.006 - Wakai, K; Azuma, K; Iwamura, C; Maimaiti, M; Mikami, K; Yoneda, K; Sakamoto, S; Ikehara, S; Yamaguchi, T; Hirahara, K; Ichikawa, T; Nakayama, T; Ikehara, Y.
The new preparation method for paraffin-embedded samples applying scanning electron microscopy revealed characteristic features in asthma-induced mice.
SCI REP. 2022 May 31;12(1):9046. doi: 10.1038/s41598-022-12666-8 - Kuroki, T; Hatta, T; Natsume, T; Sakai, N; Yagi, A; Kato, K; Nagata, K; Kawaguchi, A.
ARHGAP1 Transported with Influenza Viral Genome Ensures Integrity of Viral Particle Surface through Efficient Budozone Formation.
MBIO. 2022 Apr 27:e0072122. doi: 10.1128/mbio.00721-22 - Natsume, T.
Robotic Biology: Robotics and AI Accelerate Life Science.
CANCER SCIENCE. 113; FEB - Demachi, A; Ohte, S; Uchida, R; Shin-Ya, K; Ohshiro, T; Tomoda, H; Ikeda, H.
Discovery of prescopranone, a key intermediate in scopranone biosynthesis.
JOURNAL OF ANTIBIOTICS. 2022 Apr 20. doi: 10.1038/s41429-022-00521-x - Maeda, F; Kato, A; Takeshima, K; Shibazaki, M; Sato, R; Shibata,T; Miyake, K; Kozuka-Hata, H; Oyama, M; Shimizu, E; Imoto, S; Miyano, S; Adachi, S; Natsume, T; Takeuchi, K; Maruzuru, Y; Koyanagi, N; Jun, A; Yasushi, K.
Role of the Orphan Transporter SLC35E1 in the Nuclear Egress of Herpes Simplex Virus 1.
J VIROL. 2022 Apr 27:e0030622. doi: 10.1128/jvi.00306-22 - Mohamed, HMA; Takahashi, A; Nishijima, S; Adachi, S; Murai, I; Okamura, H; Yamamoto, T.
CNOT1 regulates circadian behaviour through Per2 mRNA decay in a deadenylation-dependent manner.
RNA BIOL. 2022;19(1):703-718. doi: 10.1080/15476286.2022.2071026 - Rider, SD Jr; Gadgil, RY; Hitch, DC; Damewood, FJ 4th; Zavada, N; Shanahan, M; Alhawach, V; Shrestha, R; Shin-Ya, K; Leffak, M.
Stable G-quadruplex DNA structures promote replication-dependent genome instability.
J BIOL CHEM. 2022 Apr 18:101947. doi: 10.1016/j.jbc.2022.101947 - Miyazaki, Y; Mori, N; Akagi, Y; Oda, T; Kida, YS.
Potential Metabolite Markers for Pancreatic Cancer Identified by Metabolomic Analysis of Induced Cancer-Associated Fibroblasts.
CANCERS. 2022 Mar 8;14(6):1375. doi: 10.3390/cancers14061375 - Ishii, N.
Systematic Investigation of Lanthanoid Transition Heavy Metal Acetates as Electron Staining Reagents for Protein Molecules in Biological Transmission Electron Microscopy.
MICROSC MICROANAL. 2022 Apr 1:1-10. doi: 10.1017/S1431927622000411 - Abdelhady, AM; Hirano, Y; Onizuka, K; Okamura, H; Komatsu, Y; Nagatsugi, F.
Synthesis of Crosslinked 2-OMe RNA Duplexes Using 2-Amino-6-Vinylpurine and Their Application for Effective Inhibition of miRNA Function.
CURR PROTOC. Mar;2(3):e386. doi: 10.1002/cpz1.386 - Nagasaki, A; Katoh, K; Hoshi, M; Doi, M; Nakamura, C; Uyeda, TQP.
Characterization of phalloidin-negative nuclear actin filaments in U2OS cells expressing cytoplasmic actin-EGFP.
GENES TO CELLS. 2022 Feb 23. doi: 10.1111/gtc.12930 - Tajima, K; Yagi, H; Morisaku, T; Nishi, K; Kushige, H; Kojima, H; Higashi, H; Kuroda, K; Kitago, M; Adachi, S; Natsume, T; Nishimura, K; Oya, M; Kitagawa, Y.
An organ-derived extracellular matrix triggers in situ kidney regeneration in a preclinical model.
NPJ REGEN MED. 2022 Feb 28;7(1):18. doi: 10.1038/s41536-022-00213-y - Nagata, R; Suemune, H; Kobayashi, M; Shinada, T; Shin-Ya, K; Nishiyama, M; Hino, T; Sato, Y; Kuzuyama, T; Nagano, S.
Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-like Enzymes.
ANGEW CHEM INT ED ENGL. 2022 Mar 2. doi: 10.1002/anie.202117430 - Mitani, Y; Yasuno, R; Kihira, K; Chung, KM; Mitsuda, N; Kanie, S; Tomioka, A; Kaji, H; Ohmiya, Y.
Host-Dependent Producibility of Recombinant Cypridina noctiluca Luciferase With Glycosylation Defects.
FRONTIERS IN BIOENGINEERING AND BIOTECHNOLOGY 10;doi: 10.3389/fbioe.2022.774786 FEB 7 - Saiki, P; Yoshihara, M; Kawano, Y; Miyazaki, H; Miyazaki, K.
Anti-Inflammatory Effects of Heliangin from Jerusalem Artichoke (Helianthus tuberosus) Leaves Might Prevent Atherosclerosis.
BIOMOLECULES 12 (1); doi:10.3390/biom12010091 JAN - Penedo, M; Miyazawa, K; Okano, N; Furusho, H; Ichikawa, T; Alam, MS; Miyata, K; Nakamura, C; Fukuma, T.
Visualizing intracellular nanostructures of living cells by nanoendoscopy-AFM.
SCI ADV. 2021 Dec 24;7(52):eabj4990. doi: 10.1126/sciadv.abj4990. Epub 2021 Dec 22 - Sogabe, M; Kojima, S; Kaya, T; Tomioka, A; Kaji, H; Sato, T; Chiba, Y; Shimizu, A; Tanaka, N; Suzuki, N; Hayashi, I; Mikami, M; Togayachi, A; Narimatsu, H.
Sensitive New Assay System for Serum Wisteria floribunda Agglutinin-Reactive Ceruloplasmin That Distinguishes Ovarian Clear Cell Carcinoma from Endometrioma.
ANALYTICAL CHEMISTRY; doi:10.1021/acs.analchem.1c04302 - Yamamoto, K; Yoneda, Y; Makino, A; Tanaka, Y; Meng, XY; Hashimoto, J; Shin-Ya, K; Satoh, N; Fujie, M; Toyama, T; Mori, K; Ike, M, Morikawa, M; Kamagata, Y; Tamaki, H.
Draft Genome Sequence of Bryobacteraceae Strain F-183.
MICROBIOL RESOUR ANNOUNC. 2022 Jan 13:e0045321. doi: 10.1128/mra.00453-21 - Hori, K; Yoshimoto, S; Yoshino, T; Zako, T; Hirao, G; Fujita, S; Nakamura, C; Yamagishi, A; Kamiya, N.
Recent advances in research on biointerfaces: From cell surfaces to artificial interfaces.
J BIOSCI BIOENG. 2022 Jan 5:S1389-1723(21)00331-5. doi: 10.1016/j.jbiosc.2021.12.004 - Ogata, H; Akita, S; Ikehara, S; Azuma, K; Yamaguchi, T; Maimaiti, M; Maezawa, Y; Kubota, Y; Yokote, K; Mitsukawa, N; Ikehara, Y.
Calcification in Werner syndrome associated with lymphatic vessels aging.
AGING (Albany NY). 2021 Dec 27;13(undefined). doi: 10.18632/aging.203789 - Matsuzawa, T; Watanabe, M; Nakamichi, Y; Kameyama, A; Kojima, N; Yaoi, K.
Characterization of an extracellular α-xylosidase involved in xyloglucan degradation in Aspergillus oryzae.
APPL MICROBIOL BIOTECHNOL. 2021 Dec 31. doi: 10.1007/s00253-021-11744-7 - Higashi, H; Yagi, H; Kuroda, K; Tajima, K; Kojima, H; Nishi, K; Morisaku, T; Hirukawa, K; Fukuda, K; Matsubara, K; Kitago, M; Shinoda, M; Obara, H; Adachi, S; Nishimura, K; Natsume, T; Tomi, M; Soto-Gutierrez, A; Kitagawa, Y.
Transplantation of Bioengineered Liver Capable of Extended Function in a Preclinical Liver Failure Model.
AM J TRANSPLANT. 2021 Dec 21. doi: 10.1111/ajt.16928 - Takayanagi, S; Watanabe, K; Maruyama, T; Ogawa, M; Morishita, K; Soga, M; Hatta, T; Natsume, T; Hirano, T; Kagechika, H; Hattori, K; Naguro, I; Ichijo, H.
ASKA technology-based pull-down method reveals a suppressive effect of ASK1 on the inflammatory NOD-RIPK2 pathway in brown adipocytes.
SCI REP. 2021 Nov 10;11(1):22009. doi: 10.1038/s41598-021-01123-7 - Adler, A; Inoue, Y; Ekdahl, KN; Baba, T; Ishihara, K; Nilsson, B; Teramura, Y.
Effect of liposome surface modification with water-soluble phospholipid polymer chain-conjugated lipids on interaction with human plasma proteins.
J MATER CHEM B. 2021 Oct 7. doi: 10.1039/d1tb01485d - Tsutsumi, H; Moriwaki, Y; Terada, T; Shimizu, K; Shin-Ya, K; Katsuyama, Y; Ohnishi, Y.
Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C-6 Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C-6 Methyltransferase.
ANGEW CHEM INT ED ENGL. 2021 Oct 9. doi: 10.1002/anie.202111217 - Takeuchi, K; Kofuku, Y; Imai, S; Ueda, T; Tokunaga, Y; Toyama, Y; Shiraishi, Y; Shimada, I.
Function-Related Dynamics in Multi-Spanning Helical Membrane Proteins Revealed by Solution NMR.
MEMBRANES (Basel). 2021 Aug 9;11(8):604. doi: 10.3390/membranes11080604 - Isaka, E; Sugiura, T; Hashimoto, K; Kikuta, K; Anazawa, U; Nomura, T; Kameyama, A.
Characterization of tumor-associated MUC1 and its glycans expressed in mucoepidermoid carcinoma.
ONCOLOGY LETTERS 22 (4);doi: 10.3892/ol.2021.12963 OCT - Perez, CAG; Adachi, S; Nong, QD; Adhitama, N; Matsuura, T; Natsume, T; Wada, T; Kato, Y; Watanabe, H.
Sense-overlapping lncRNA as a decoy of translational repressor protein for dimorphic gene expression.
PLOS GENET. 2021 Jul 28;17(7):e1009683. doi: 10.1371/journal.pgen.1009683 - Konno, S; Kobayashi, K; Senda, M; Funai, Y; Seki, Y; Tamai, I; Schäkel, L; Sakata, K; Pillaiyar, T; Taguchi, A; Taniguchi, A; Gütschow, M; Müller, CE; Takeuchi, K; Hirohama, M; Kawaguch,i A; Kojima, M; Senda, T; Shirasaka, Y; Kamitani, W; Hayashi, Y.
3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J MED CHEM. 2021 Jul 27. doi: 10.1021/acs.jmedchem.1c00665 - Haramoto, Y.
First person - Yoshikazu Haramoto.
BIOLOGY OPEN 10 (4);doi: 10.1242/bio.058759 APR - Seimiya, H; Nagasawa, K; Shin-Ya, K.
Chemical targeting of G-quadruplexes in telomeres and beyond for molecular cancer therapeutics.
J ANTIBIOT (Tokyo). 2021 Jul 20. doi: 10.1038/s41429-021-00454-x - Mostafa, Abdelhady A; Hirano, Y; Onizuka, K; Okamura, H; Komatsu, Y; Nagatsugi, F.
Synthesis of Crosslinked 2-OMe RNA Duplexes and Their Application for Effective Inhibition of miRNA Function.
BIOORG MED CHEM LETT. 2021 Jul 8:128257. doi: 10.1016/j.bmcl.2021.128257 - Ninomiya, K; Iwakiri, J; Aly, MK; Sakaguchi, Y; Adachi, S; Natsume, T; Terai, G; Asai, K; Suzuki, T; Hirose, T.
m(6) A modification of HSATIII lncRNAs regulates temperature-dependent splicing.
EMBO J. 2021 Jun 29:e107976. doi: 10.15252/embj.2021107976 - Okumura, S; Hirano, Y; Komatsu, Y.
Stable duplex-linked antisense targeting miR-148a inhibits breast cancer cell proliferation.
SCI REP. 2021 Jun 1;11(1):11467. doi: 10.1038/s41598-021-90972-3 - Suzuki, T; Katayama, Y; Komatsu, Y; Kamiya, H.
Similar frequency and signature of untargeted substitutions induced by abasic site analog under reduced human APE1 conditions.
J TOXICOL SCI. 2021;46(6):283-288. doi: 10.2131/jts.46.283 - Yoneda, Y; Yamamoto, K; Makino, A; Tanaka, Y; Meng, XY; Hashimoto, J; Shin-Ya, K; Satoh, N; Fujie, M; Toyama, T; Mori, K; Ike, M; Morikawa, M; Kamagata, Y; Tamaki, H
Novel Plant-Associated Acidobacteria Promotes Growth of Common Floating Aquatic Plants, Duckweeds
Microorganisms. 2021 May 24;9(6):1133. doi: 10.3390/microorganisms9061133 - Mogi, K; Kimura, H; Kondo, Y; Inoue, T; Adachi, S; Natsume, T.
Automatic radioisotope manipulation for small amount of nuclear medicine using an EWOD device with a dimple structure.
R SOC OPEN SCI. 2021 May 26;8(5):201809. doi: 10.1098/rsos.201809 - Amemiya, T; Horimoto, K; Fukui, K.
Application of multiple omics and network projection analyses to drug repositioning for pathogenic mosquito-borne viruses.
SCI REP. 2021 May 12;11(1):10136. doi: 10.1038/s41598-021-89171-x - Baba, T; Tanimura, S; Yamaguchi, A; Horikawa, K; Yokozeki, M; Hachiya, S; Iemura, SI; Natsume, T; Matsuda, N; Takeda, K.
Cleaved PGAM5 dephosphorylates nuclear serine/arginine-rich proteinsduring mitophagy.
BIOCHIM BIOPHYS ACTA MOL CELL RES. 2021 Apr 16:119045. doi: 10.1016/j.bbamcr.2021.119045 - Akagi, Y; Mori, N; Kawamura, T; Takayama, Y; Kida, YS.
Non-invasive cell classification using the Paint Raman Express Spectroscopy System (PRESS)
SCI REP. 2021 Apr 23;11(1):8818. doi: 10.1038/s41598-021-88056-3 - Kaneko, S; Mitsuyama, T; Shiraishi, K; Ikawa, N; Shozu, K; Dozen, A; Machino, H; Asada, K; Komatsu, M; Kukita, A; Sone, K; Yoshida, H; Motoi, N; Hayami, S; Yoneoka, Y; Kato, T; Kohno, T; Natsume, T; Keudell, GV; Saloura, V; Yamaue, H; Hamamoto, R
Genome-Wide Chromatin Analysis of FFPE Tissues Using a Dual-Arm Robot with Clinical Potential
Cancers (Basel). 2021 Apr 28;13(9):2126. doi: 10.3390/cancers13092126 - Kohrogi, K; Hino, S; Sakamoto, A; Anan, K; Takase, R; Araki, H; Hino, Y; Araki, K; Sato, T; Nakamura, K; Nakao, M
LSD1 defines erythroleukemia metabolism by controlling the lineage-specific transcription factors GATA1 and C/EBP
Blood Adv. 2021 May 11;5(9):2305-2318. doi: 10.1182/bloodadvances.2020003521 - Okubo, S; Ena, E; Okuda, A; Kozone, I; Hashimoto, J; Nishitsuji, Y; Fujie, M; Satoh, N; Ikeda, H; Shin-Ya, K.
Identification of functional cytochrome P450 and ferredoxin from Streptomyces sp. EAS-AB2608 by transcriptional analysis and their heterologous expression.
APPL MICROBIOL BIOTECHNOL. 2021 May 4. doi: 10.1007/s00253-021-11304-z - Mori, N; Kida, YS.
Applicability of Artificial Vascularized Liver Tissue to Proteomic Analysis.
MICROMACHINES (Basel). 2021 Apr 11;12(4):418. doi: 10.3390/mi12040418 - Penedo, M; Shirokawa, T; Alam, MS; Miyazawa, K; Ichikawa, T; Okano, N; Furusho, H; Nakamura, C; Fukuma, T.
Cell penetration efficiency analysis of different atomic force microscopy nanoneedles into living cells.
SCI REP. 2021 Apr 8;11(1):7756. doi: 10.1038/s41598-021-87319-3 - Ohashi, H; Watashi, K; Saso, W; Shionoya, K; Iwanami, S; Hirokawa, T; Shirai, T; Kanaya, S; Ito, Y; Kim, KS; Nomura, T; Suzuki, T; Nishioka, K; Ando, S; Ejima, K; Koizumi, Y; Tanaka, T; Aoki, S; Kuramochi, K; Suzuki, T; Hashiguchi, T; Maenaka, K; Matano, T; Muramatsu, M; Saijo, M; Aihara, K; Iwami, S; Takeda, M; McKeating, JA; Wakita, T
Potential anti-COVID-19 agents, Cepharanthine and Nelfinavir, and their usage for combination treatment.
ISCIENCE. 2021 Mar 26:102367. doi: 10.1016/j.isci.2021.102367 - Hirama, H; Otahara, R; Mogi, K; Hayase, M; Torii, T; Mekaru, H.
Rapid Prototyping of a Nanoparticle Concentrator Using a Hydrogel Molding Method.
POLYMERS (Basel). 2021 Mar 29;13(7):1069. doi: 10.3390/polym13071069 - Katsuyama, Y; Sone, K; Harada, A; Kawai, S; Urano, N; Adachi, N; Moriya, T; Kawasaki, M; Shin-Ya, K; Senda, T; Ohnishi, Y
Structural and functional analyses of the tridomain-nonribosomal peptide synthetase FmoA3 for 4-methyloxazoline ring formation
Angew Chem Int Ed Engl. 2021 Mar 29. doi: 10.1002/anie.202102760 - Nakamura, A; Wang, D; Komatsu, Y.
Analysis of GTP addition in the reverse (3-5) direction by human tRNA(His) guanylyltransferase.
RNA. 2021 Mar 23:rna.078287.120. doi: 10.1261/rna.078287.120