Simultaneous decoding of 10,000 cells’ glycome and transcriptome in single cells
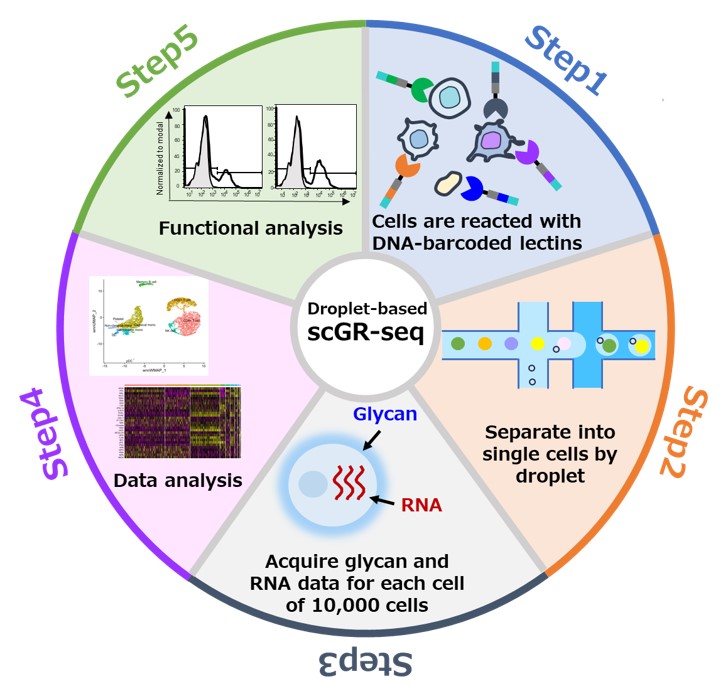
Hiroaki Tateno, a research group leader, and Sunanda Keisham, a Ph.D student in the Human Biology Program at University of Tsukuba, have developed a droplet-type single-cell glycan-RNA sequencing method (droplet-type scGR-seq method) that simultaneously analyzes glycans and RNAs expressed on each of approximately 10,000 cells isolated from tissues and other sources using a next-generation sequencer. Cell surface glycans are used as drug targets for various diseases and for quality control of cells for regenerative medicine. Previously, we developed a technology to obtain information on the expression of extracellular glycans and intracellular RNA in each cell using a next-generation sequencer after reacting multiple cells with lectins (glycan-binding proteins) labeled with DNA barcodes and separating each cell into a microcentrifuge tube. However, the number of cells that can be analyzed in a single experiment is limited to several hundred by the conventional plate-type method in which each cell is dispensed into a microcentrifuge tube. With the introduction of droplet technology, approximately 10,000 cells can be analyzed in a single experiment, a 100-fold increase in the number of cells that can be processed compared to the conventional method. This technology enables more comprehensive analysis of the differences between different cell types and the diversity of cell surface sugar chains within the same cell type, as well as rapid acquisition of information on rare cells that contain only a small number of glycans.Details of this technology were published online in Small Methods on January 2, 2024.
※Please see the press release below the image for details.
Press release
Publication
- Journal:Small Methods January 2, 2024.
- Title:Droplet-Based Glycan and RNA Sequencing for Profiling the Distinct Cellular Glyco-States in Single Cells
- Authors:Sunanda Keisham, Sayoko Saito, Satori Kowashi, Hiroaki Tateno