Biological Data Science Research Group
Biological Data Science Group Research Group Overview
One of the defining features of our research group is the seamless integration of wet and dry research. We not only develop innovative in silico technologies to support drug discovery and diagnostics but also harness the synergy between experimental and computational approaches. By combining advanced measurement techniques and novel production methods with cutting-edge computational modeling and data analysis, we elevate our technologies to new heights. Our mission is to accelerate breakthroughs in drug discovery, diagnostics, and treatment technologies. Furthermore, we are fostering the next generation of researchers who excel in both wet and dry research domains.
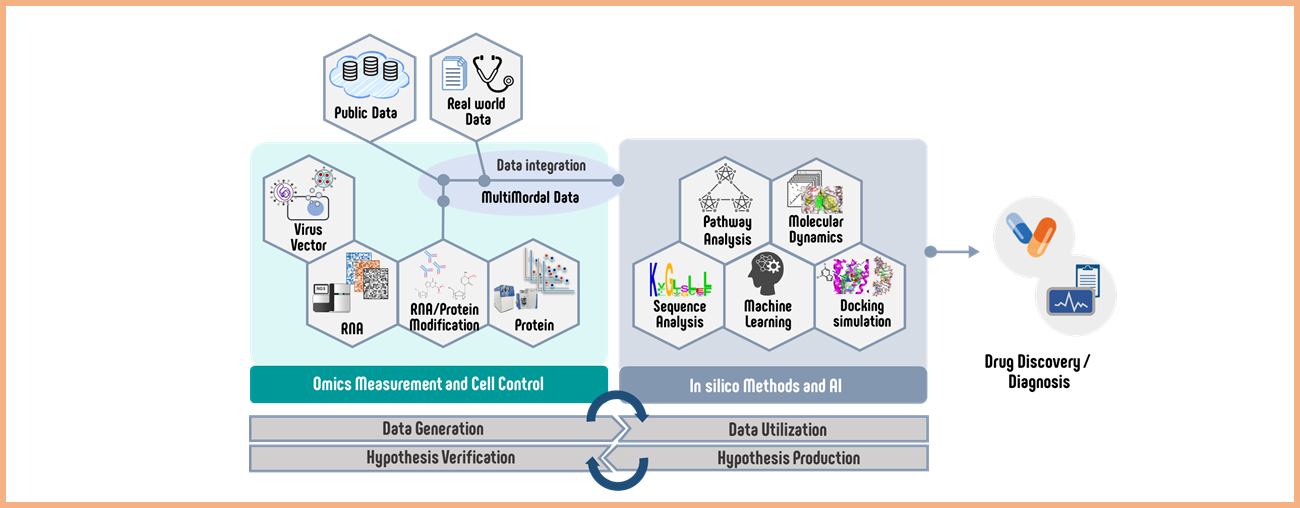
Research Project
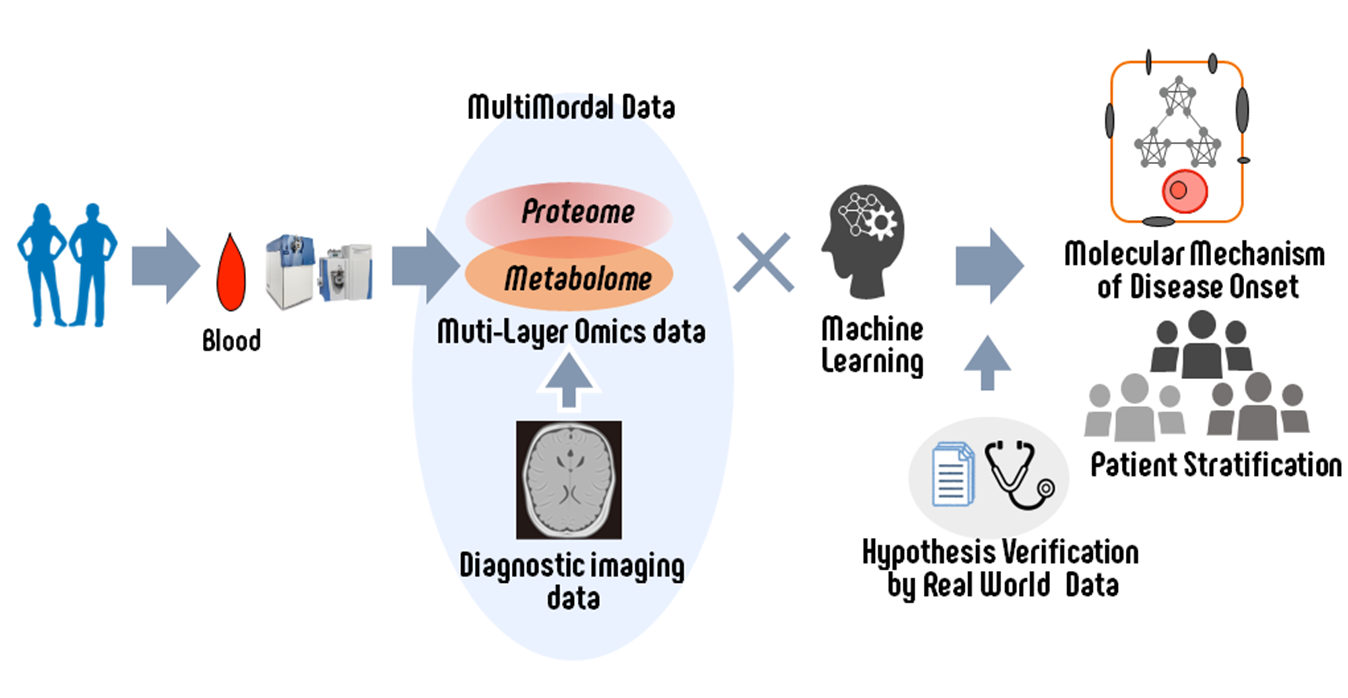
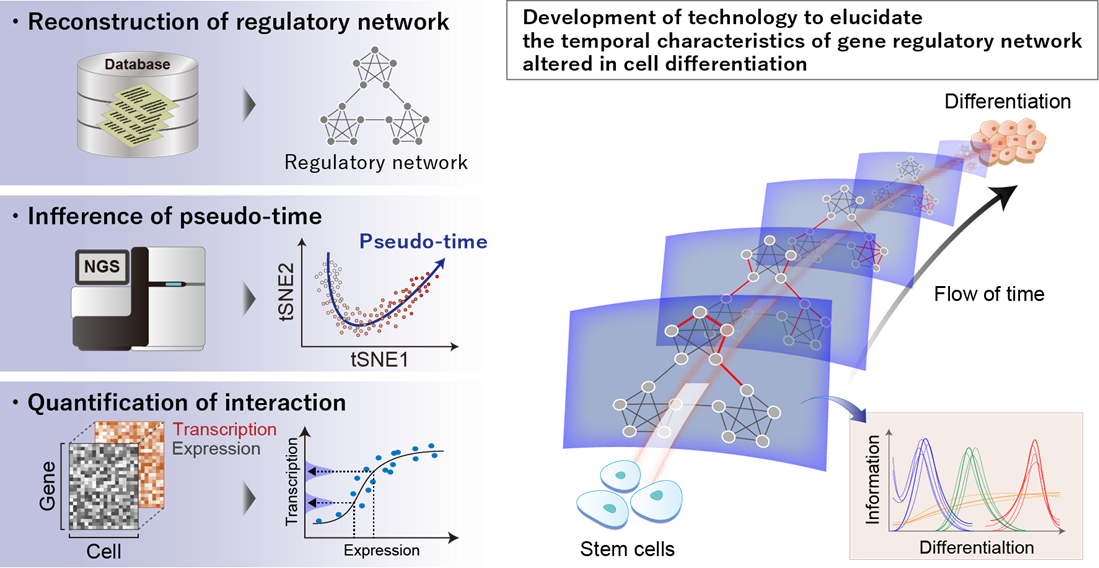
project 1:Biological data science based on multimodal data analysis
Researcher:IMAI Kenichiro (Internal collaboration within the research group: KAGIWADA Harumi, KONNO Masamitsu)
We are conducting multimodal data analysis by integrating multi-layer omics data with analytical data such as diagnostic imaging. Through this approach, we aim to elucidate the molecular mechanisms involved in disease onset and develop technologies that contribute to early diagnosis. In addition, we are utilizing real-world data, such as health insurance claim (receipt) data, to validate the disease mechanisms identified through our analyses.
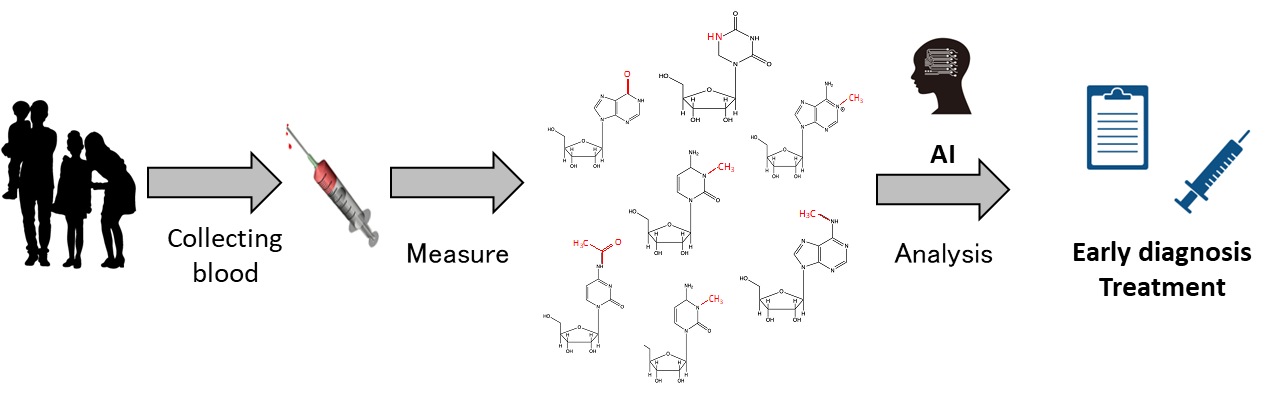
Project 2:Development of new diagnosis and treatment of diseases using omics modification
Researcher: KONNO Masamitsu (Internal collaboration within the research group: KOSEKI Jun, IMAI Kenichiro)
RNA modifications are known to be changed in various diseases. However, the measurement technology is still in development, and the application to diagnosis and treatment has not yet been realized. Therefore, we are developing a new technology using AI to measure modification information of omics, especially RNA. We are also analyzing the modification information to develop early diagnosis technology and new treatment methods for various diseases, including cancer.
Project 3: Development of drug discovery support technology based on theoretical structural analysis
Researcher:KOSEKI Jun (Internal collaboration within the research group: MOTONO Chie, IMAI Kenichiro)
It is well known that changes in a protein’s “structure” can affect its “function”. However, for proteins whose structures remain unknown, it has been difficult to accurately understand the relationship between structure and function. Recently, AI-based techniques have made it possible to predict the three-dimensional structure of proteins with high accuracy. However, identifying key functional sites and predicting how amino acid mutations affect function remains a challenge. Therefore, we have developed a theoretical structural analysis method to address this issue. By identifying hidden functional sites in undruggable proteins, we aim to support drug discovery through the design of small to medium-sized molecules and even binding proteins.
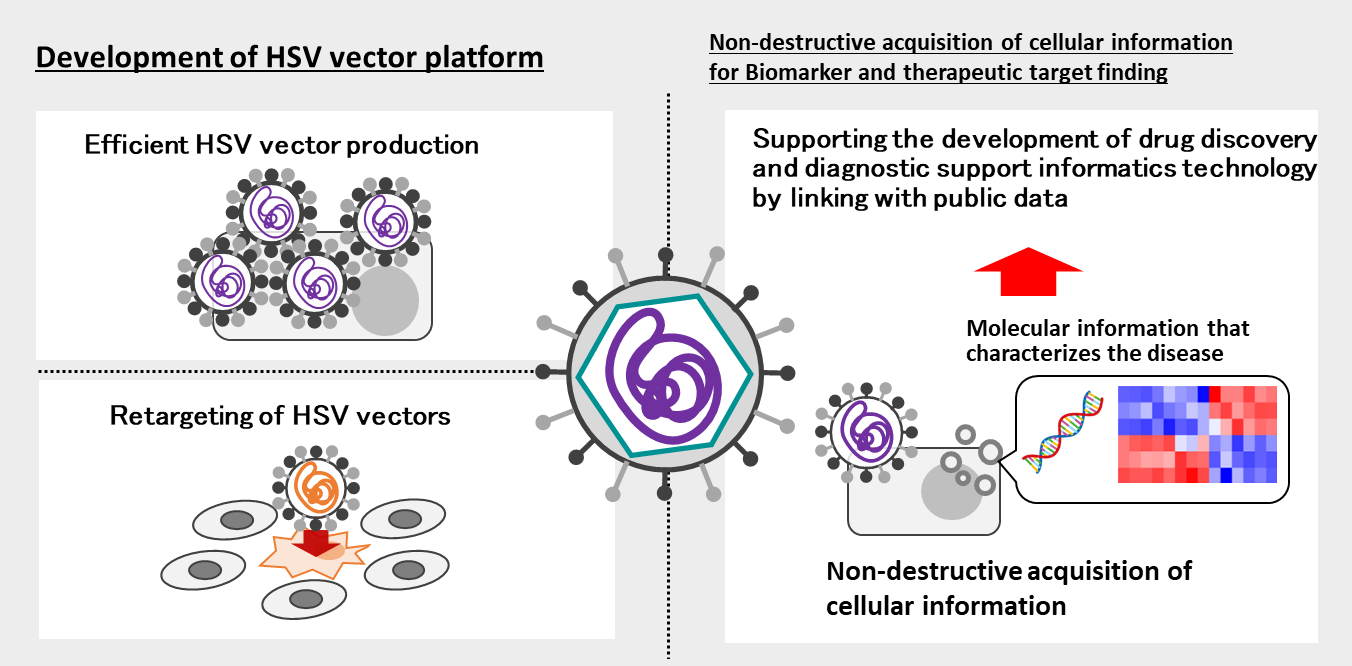
Project 4:Development of herpesvirus vectors as a drug modality
Researcher:MAEDA Fumio (Internal collaboration within the research group: KOSEKI Jun, IMAI Kenichiro)
The herpes simplex virus Amplicon vector eliminates the viral genome from the viral particle and packages a plasmid instead, and is a viral vector characterized by (1) the limited cytotoxicity, (2) the large transgene capacity of up to 150 kb, and (3) the broad host range of vector transduction. We are currently developing methods for efficient production of HSV vectors, retargeting them, and utilizing HSV vectors as a gene therapy drug.
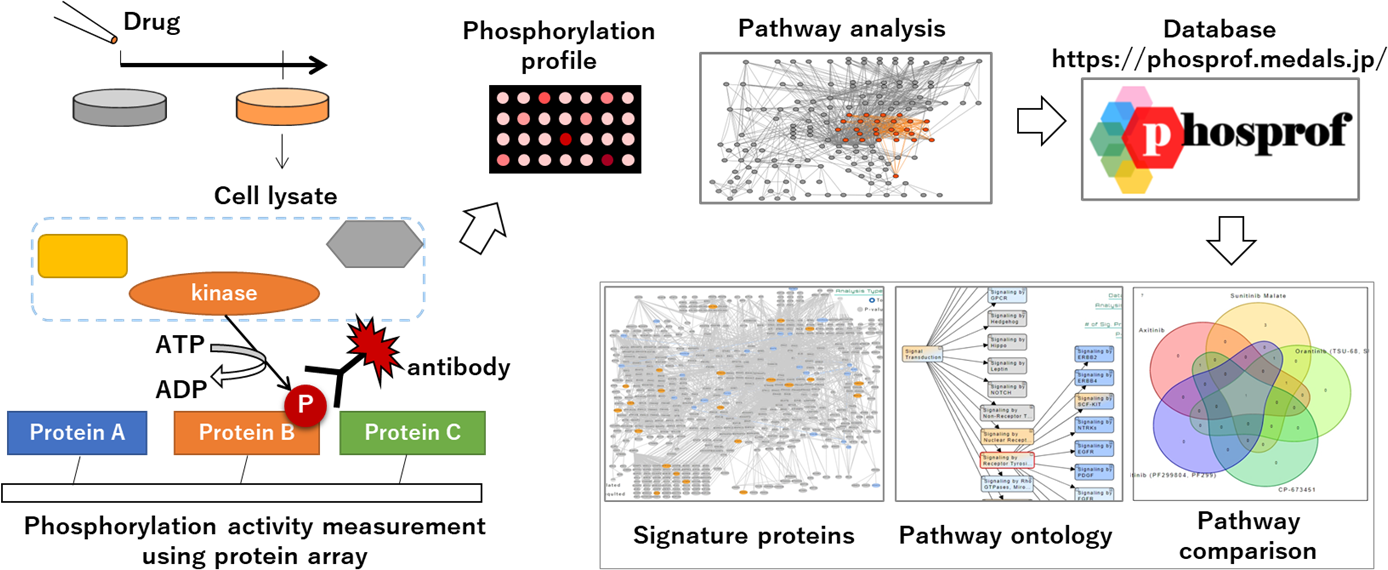
Project 5:Protein network dynamics analysis
Researcher: KAGIWADA Harumi (Internal collaboration within the research group: MAEDA Fumio, IMAI Kenichiro)
Protein phosphorylation is one of the representative mechanisms of post-translational modification which regulates enzyme activities and transduces signaling pathways. Here we developed a novel and convenient system for a series of phosphorylation analyses. By our system, the active pathways can be visualized on the pathway map to facilitate biological interpretation depending on the different cases such as disease-control and drug treatment. Pathway analysis results of drug response using this system have been collected and published as Phosprof database.
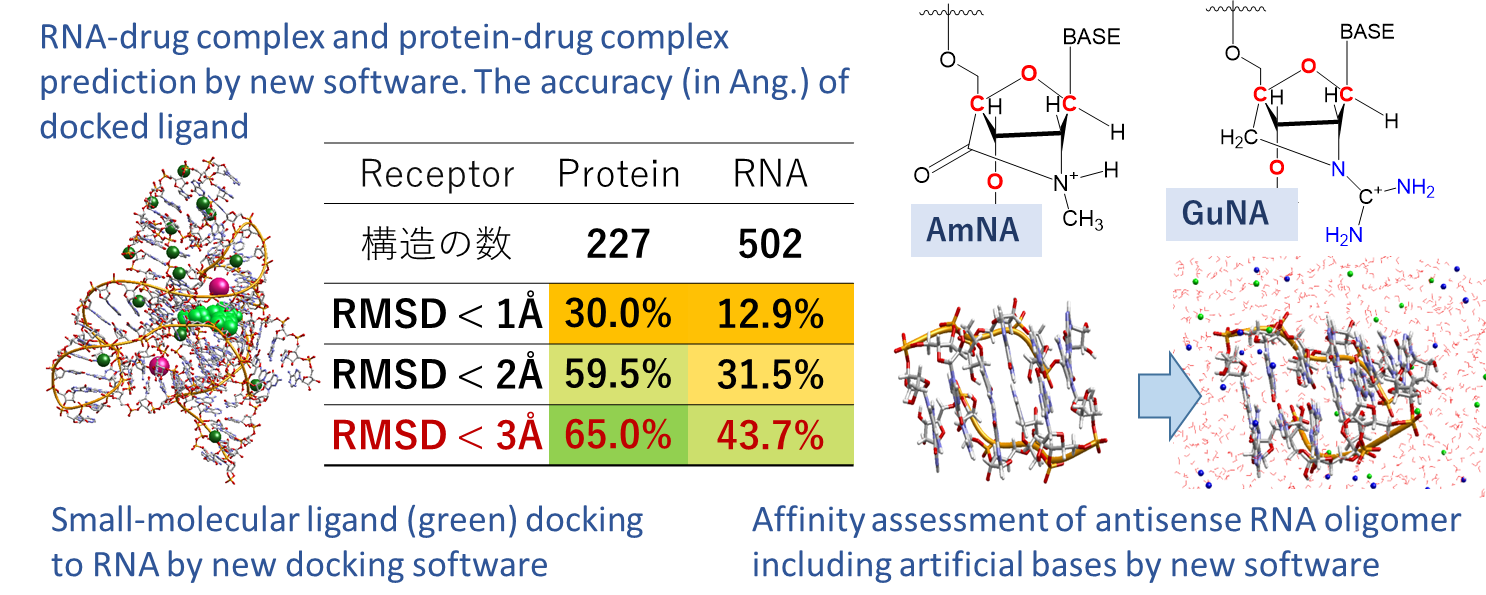
Project6: Development of RNA simulation technology for new drugs and diagnosis
Researcher: FUKUNISHI Yoshifumi (Internal collaboration within the research group: MOTONO Chie, IMAI Kenichiro)
Since the COVID-19 vaccine, RNA has been expanding as a drug molecule (modality), drug target, and diagnosis marker, but as various side effects show, there are many unknown properties, and it is thought that it will be 5~10 years before it will be widely used. RNA exerts its function through molecular motion in addition to sequence information and 3D structure, but due to the difficulty of experiments, the structural information is only 1/300 of that of proteins. It is a difficult theme for AI, which learns large amounts of data and exerts its predictive power. We have developed a method to compare the theoretical calculation accuracy of RNAs with experiments and are optimizing the calculation method. We will implement the calculation technology that we have cultivated in collaboration with >40 pharmaceutical and IT companies in Japan into the drug discovery software myPresto, and develop and provide theoretical tools to elucidate the movement of biomolecules that regulate life activities, molecular recognition, and control mechanisms such as base modification.
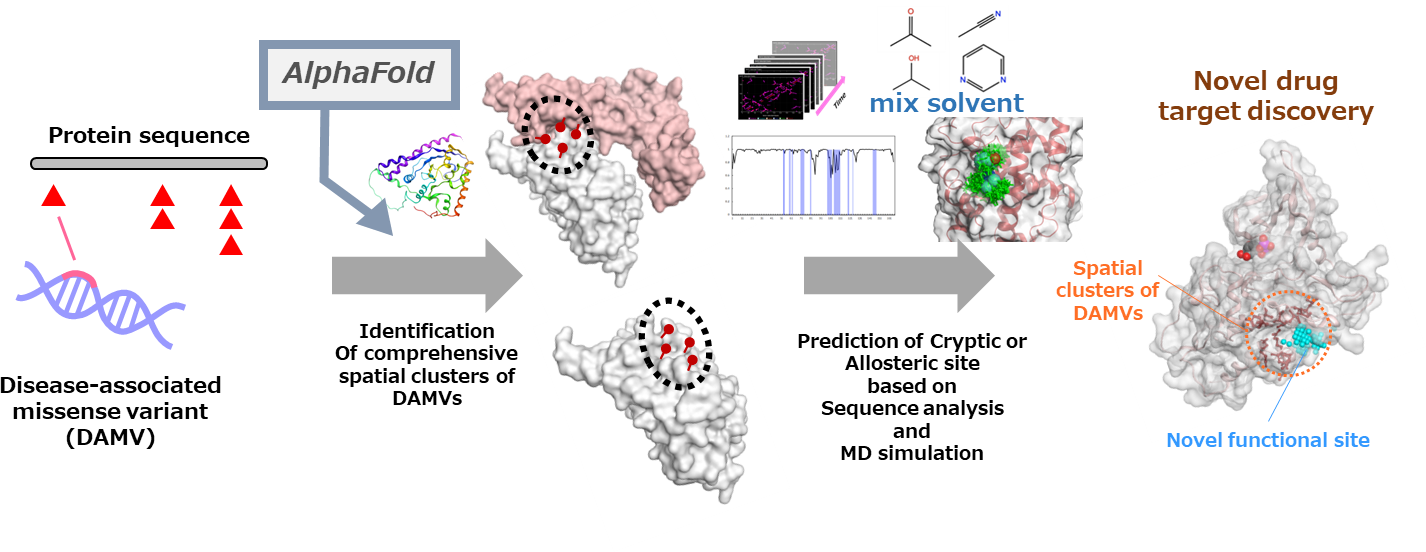
Project7: Search for undiscovered protein functional sites based on the spatial distribution of disease-associated missense variants
Researcher: MOTONO Chie, IMAI Kenichiro
Effective utilization of large number of disease-associated variants revealed through sequencing projects is one of the major challenges in medical and pharmaceutical field. Disease-associated missense variants (DAMVs) tend to gather around functional sites such as ligand binding sites and PPI interfaces. In this study, we comprehensively defined 3D variant clusters by clustering of the spatially-distributed DAMVs on human porotein structures utilizing AlphaFold2 predicted structures. The 3D variant clusters not related to known functional sites would indicate undiscovered functional sites such as cryptic sites or allosteric sites. We are developing method for prediction of novel functional sites leading to novel drug targets based on the 3D variant clusters using mixed solvent molecular dynamics simulation.
Member
| photo | position & name | field of expertise | and other info |
|---|---|---|---|
 |
Research Group Leader(Attached to Research Group) IMAI Kenichiro |
|
|
 |
Senior researcher KONNO Masamitsu |
|
|
 |
Senior Researcher KOSEKI Jun |
|
|
 |
Senior researcher KAGIWADA Harumi |
|
|
 |
Researcher MAEDA Fumio |
|
|
 |
Attached to Research Group FUKUNISHI Yoshifumi |
|
|
 |
Attached to Research Group MOTONO Chie |
|
Results
- Bekker, GJ; Fukunishi, Y; Higo, J; Kamiya, N.
Assessment of RNA Force Fields for Dynamic Docking of Small Molecules Using Multicanonical MD Simulations.
JOURNAL OF CHEMICAL THEORY AND COMPUTATION. 2025 Oct 22. doi:10.1021/acs.jctc.5c01114 - Ojima-Kato, T; Yokoyama, G; Nakano, H; Hamada, M; Motono, C.
Screening and machine learning-based prediction of translation-enhancing peptides that reduce ribosomal stalling in Escherichia coli.
RSC CHEM BIOL. 2025 Oct 22. doi:10.1039/d5cb00199d - Hara, T; Meng, S; Motooka, D; Arao, Y; Saito, Yk Rennie, S; Uchida, S; Ofusa, K; Arai, T; Konno, M; Satoh, T; Ishii, H.
Deficiencies in methionine, tryptophan, and niacin remodels intestinal transcriptome and gut microbiota in female mice.
SCI REP. 2025 Oct 16;15(1):36155. doi:10.1038/s41598-025-18046-2 - Lintuluoto, M; Horioka, Y; Fujiwara, M; Abe, M; Fukunishi, Y; Tamura, H; Lintuluoto, JM.
Investigation on the Substrate Specificity of Serine Protease Neuropsin by Molecular Dynamics Simulation and Marmoset Gene Atlas (MGA.
ACS OMEG. 2025 JUN 2. doi:10.1021/acsomega.5c00843 - Motono, C; Yanagisawa, K; Koseki, J; Imai, K.
CrypTothML: An Integrated Mixed-Solvent Molecular Dynamics Simulation and Machine Learning Approach for Cryptic Site Prediction.
INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2025 May 14;26(10):4710. doi:10.3390/ijms26104710 - Koseki, J; Motono, C; Yanagisawa, K; Kudo, G; Yoshino, R; Hirokawa, T; Imai, K.
CrypToth: Cryptic Pocket Detection through Mixed-Solvent Molecular Dynamics Simulations-Based Topological Data Analysis.
JOURNAL OF CHEMICAL INFORMATION AND MODELING. 2025 May 22. doi:10.1021/acs.jcim.4c02111 - Nobe, M; Maruzuru, Y; Takeshima, K; Maeda, F; Kusano, H; Yoshimura, R; Nishiyama, T; Park, H; Kozaki, Y; Iwami, S; Koyanagi, N; Kato, A; Natsume, T; Adachi, S; Kawaguchi, Y.
Direct relationship between protein expression and progeny yield of herpes simplex virus 1.
MBIO. 2025 May 5:e0028025. doi:10.1128/mbio.00280-25 - Ohkawa, M; Ohshiro, T; Koseki, J; Kon, S; Ozeki, Y; Taniguchi, M; Shimamura, T; Konno, M.
New RNA modifications induce malignancy for pancreatic cancer.
CANCER SCIENCE. 2025 Mar. doi: 10.1111/cas.16114 - Yamagishi, A; Tokuoka, R; Imai, K; Mizusawa, M; Susaki, M; Uchida, K; Kijima, ST; Nagasaki, A; Takeshita, D; Yoshikawa, C; Uyeda, TQP; Nakamura, C.
Nestin Forms a Flexible Cytoskeleton by Means of a Huge Tail Domain That Is Reversibly Stretched and Contracted by Weak Forces.
CELLS. 2025 Jan 17;14(2):138. doi:10.3390/cells14020138 - Sato, T; Abe, K; Koseki, J; Seto, M; Yokoyama, J; Akashi, T; Terada, M; Kadowaki, K; Yoshida, S; Yamashiki, YA; Shimamura, T.
Survivability and life support in sealed mini-ecosystems with simulated planetary soils.
SCI REP. 2024 Nov 1;14(1):26322. doi: 10.1038/s41598-024-75328-x - Yoshinori, F; Imai, K; Horton, P.
Prediction of mitochondrial targeting signals and their cleavage sites.
METHODS ENZYMOL. 2024 Sep 11. doi: 10.1016/bs.mie.2024.07.026 - Fujii, Y; Kamata, K; Gerdol, M; Hasan, I; Rajia, S; Kawsar, SMA; Padma, S; Chatterjee, BP; Ohkawa, M; Ishiwata, R; Yoshimoto, S; Yamada, M; Matsuzaki, N; Yamamoto, K; Niimi, Y; Miyanishi, N; Konno, M; Pallavicini, A; Kawasaki, T; Ogawa, Y; Ozeki, Y; Fujita, H.
Multifunctional Cell Regulation Activities of the Mussel Lectin SeviL: Induction of Macrophage Polarization toward the M1 Functional Phenotype.
MARINE DRUGS. 2024 Jun 11;22(6):269. doi: 10.3390/md22060269 - Watanabe, A; Koyano, F; Imai, K; Hizukuri, Y; Ogiwara, S; Ito, T; Miyamoto, J; Shibuya, C; Kimura, M; Toriumi, K; Motono, C; Arai, M; Tanaka, K; Akiyama, Y; Yamano, K; Matsuda, N.
The origin of esterase activity of Parkinsons disease causative factor DJ-1 implied by evolutionary trace analysis of its prokaryotic homolog HchA.
J BIOL CHEM. 2024 Jun 13:107476. doi: 10.1016/j.jbc.2024.107476 - Higo, J; Bekker, GJ; Kamiya, N; Fukuda, I; Fukunishi, Y;.
Binding free-energy landscapes of small molecule binder and non-binder to FMN riboswitch: All-atom molecular dynamics.
BIOPHYS PHYSICOBIOL. 2023 Dec 13;20(4):e200047. doi: 10.2142/biophysico.bppb-v20.0047 - Bekker, GJ; Fukunishi, Y; Higo, J; Kamiya, N.
Binding Mechanism of Riboswitch to Natural Ligand Elucidated by McMD-Based Dynamic Docking Simulations.
ACS OMEGA. 2024 Jan 10;9(3):3412-3422. doi: 10.1021/acsomega.3c06826 eCollection - Germany, EM; Thewasano, N; Imai, K; Maruno, Y; Bamert, RS; Stubenrauch, CJ; Dunstan, RA; Ding, Y; Nakajima, Y; Lai, X; Webb, CT; Hidaka, K; Tan, KS; Shen, H; Lithgow, T; Shiota, T.
Dual recognition of multiple signals in bacterial outer membrane proteins enhances assembly and maintains membrane integrity.
ELIFE. 2024 Jan 16;12:RP90274. doi: 10.7554/eLife.90274 - Shimizu, R; Murai, K; Tanaka, K; Sato, Y; Takeda, N; Nakasyo, S; Shirasaki, T; Kawaguchi, K; Shimakami, T; Nio, K; Nakaya, Y; Kagiwada, H Horimoto, K; Mizokami, M; Kaneko, S; Murata, K; Yamashita, T; Honda, M.
Nucleos(t)ide analogs for hepatitis B virus infection differentially regulate the growth factor signaling in hepatocytes.
HEPATOL COMMUN. 2024 Jan 5;8(1):e0351. doi: 10.1097/HC9.0000000000000351 - Fukuda, I; Moritsugu, K; Higo, J; Fukunishi, Y.
A cutoff-based method with charge-distribution-data driven pair potentials for efficiently estimating electrostatic interactions in molecular systems.
J CHEM PHYS. 2023 Dec 21;159(23):234116. doi: 10.1063/5.0172270 - Watanabe, Y; Iwasaki, Y; Sasaki, K; Motono, C; Imai, K; Suzuki, K.
Atg15 is a vacuolar phospholipase that disintegrates organelle membranes.
CELL REP. 2023 Dec 15:113567. doi: 10.1016/j.celrep.2023.113567 - Maeda, F; Adachi, S; Natsume, T.
Non-destructive and efficient method for obtaining miRNA information in cells by artificial extracellular vesicles.
SCI REP. 2023 Dec 14;13(1):22231. doi: 10.1038/s41598-023-48995-5 - Ohkawa, M; Konno, M.
RNA Modification Related Diseases and Sensing Methods.
APPLIED SCIENCES-BASEL. 2023 13(11) 6376. doi: 10.3390/app13116376 - Shin, WH; Kumazawa, K; Imai, K; Hirokawa, T; Kihara, D.
Quantitative comparison of protein-protein interaction interface using physicochemical feature-based descriptors of surface patches.
FRONT MOL BIOSCI. 2023 Feb 6;10:1110567. doi: 10.3389/fmolb.2023.1110567 - Takeda, H; Busto, JV; Lindau, C; Tsutsumi, A; Tomii, K; Imai, K; Yamamori, Y; Hirokawa, T; Motono, C; Ganesan, I; Wenz, LS; Becker, T; Kikkawa, M; Pfanner, N; Wiedemann, N; Endo, T.
A multipoint guidance mechanism for β-barrel folding on the SAM complex.
NAT STRUCT MOL BIOL. 2023 Jan 5. doi: 10.1038/s41594-022-00897-2 - Hayashida, R; Kikuchi, R; Imai, K; Kojima, W; Yamada, T; Iijima, M; Sesaki, H; Tanaka, K; Matsuda, N; Yamano, K.
Elucidation of ubiquitin-conjugating enzymes that interact with RBR-type ubiquitin ligases using a liquid-liquid phase separation-based method.
J BIOL CHEM. 2022 Dec 20:102822. doi: 10.1016/j.jbc.2022.102822 - Murotomi, K; Kagiwada, H; Hirano, K; Yamamoto, S; Numata, N; Matsumoto, Y; Kaneko, H; Namihira, M.
Cyclo-glycylproline attenuates hydrogen peroxide-induced cellular damage mediated by the MDM2-p53 pathway in human neural stem cells.
J CELL PHYSIOL. 2022 Dec 31. doi: 10.1002/jcp.30940 - Fukunishi, Y; Higo, J; Kasahara, K.
Computer simulation of molecular recognition in biomolecular system: from in silico screening to generalized ensembles.
BIOPHYS REV. 2022 Nov 28:1-25. doi: 10.1007/s12551-022-01015-8 - Lintuluoto, M; Abe, M; Horioka, Y; Fukunishi, Y; Tamura, H; M, Lintuluoto J.
Investigation on substrate specificity and catalytic activity of serine protease neuropsin.
BIOPHYS PHYSICOBIOL. 2022 Sep 22;19:e190040. doi: 10.2142/biophysico.bppb-v19.0040 - Kagiwada, H; Motono, C; Horimoto, K; Fukui, K.
Phosprof: pathway analysis database of drug response based on phosphorylation activity measurements.
DATABASE (Oxford). 2022 Aug 22;2022:baac072. doi: 10.1093/database/baac072 - Higo, J; Kasahara, K; Bekker, GJ; Ma, B; Sakuraba, S; Iida, S; Kamiya, N; Fukuda, I; Kono, H; Fukunishi, Y; Nakamura, H.
Fly casting with ligand sliding and orientational selection supporting complex formation of a GPCR and a middle sized flexible molecule.
SCI REP. 2022 Aug 13;12(1):13792. doi: 10.1038/s41598-022-17920-7 - Hirata, Y; Oda, AH; Motono, C; Shiro, M; Ohta, K.
Imputation-free reconstructions of three-dimensional chromosome architectures in human diploid single-cells using allele-specified contacts.
SCI REP. 2022 Jul 11;12(1):11757. doi: 10.1038/s41598-022-15038-4 - Kose, S; Imai, K; Watanabe, A; Nakai, A; Suzuki, Y; Imamoto, N.
Lack of Hikeshi activates HSF1 activity under normal conditions and disturbs the heat-shock response.
LIFE SCI ALLIANCE. 2022 May 17;5(9):e202101241. doi: 10.26508/lsa.202101241 - Maeda, F; Kato, A; Takeshima, K; Shibazaki, M; Sato, R; Shibata,T; Miyake, K; Kozuka-Hata, H; Oyama, M; Shimizu, E; Imoto, S; Miyano, S; Adachi, S; Natsume, T; Takeuchi, K; Maruzuru, Y; Koyanagi, N; Jun, A; Yasushi, K.
Role of the Orphan Transporter SLC35E1 in the Nuclear Egress of Herpes Simplex Virus 1.
J VIROL. 2022 Apr 27:e0030622. doi: 10.1128/jvi.00306-22 - Santos, HJ; Hanadate, Y; Imai, K; Watanabe, H; Nozaki, T.
Entamoeba histolytica EHD1 Is Involved in Mitosome-Endosome Contact.
MBIO. 2022 Apr 11:e0384921. doi: 10.1128/mbio.03849-21 - Araiso, Y; Imai, K; Endo, T.
Role of the TOM Complex in Protein Import into Mitochondria: Structural Views.
ANNU REV BIOCHEM. 2022 Feb 14. doi: 10.1146/annurev-biochem-032620-104527 - Ono, J; Koshimizu, U; Fukunishi, Y; Nakai, H.
Multiple protonation states in ligand-free SARS-CoV-2 main protease revealed by large-scale quantum molecular dynamics simulations.
CHEM PHYS LETT. 2022 Feb 22;794:139489. doi: 10.1016/j.cplett.2022.139489 - Queliconi BB, Kojima W, Kimura M, Imai K, Udagawa C, Motono C, Hirokawa T, Tashiro S, Caaveiro JMM, Tsumoto K, Yamano K, Tanaka K, Matsuda N.
Unfolding is the driving force for mitochondrial import and degradation of Parkinsons disease-related protein DJ-1.
J CELL SCI. 2021 Oct 22:jcs.258653. doi: 10.1242/jcs.258653 - Kimura, M; Imai, K; Morinaka, Y; Hosono-Sakuma, Y; Horton, P; Imamoto, N.
Distinct mutations in importin-β family nucleocytoplasmic transport receptors transportin-SR and importin-13 affect specific cargo binding.
SCI REP. 2021 Aug 2;11(1):15649. doi: 10.1038/s41598-021-94948-1 - Motono, C; Yanagida, S; Sato, M; Hirokawa, T.
MDContactCom: a tool to identify differences of protein molecular dynamics from two MD simulation trajectories in terms of interresidue contacts.
BIOINFORMATICS. 2021 Jul 21:btab538. doi: 10.1093/bioinformatics/btab538 - Iida, S; Fukunishi, Y.
Asymmetric dynamics of dimeric SARS-CoV-2 and SARS-CoV main proteases in an apo form: Molecular dynamics study on fluctuations of active site, catalytic dyad, and hydration water.
BBA ADV. 2021;1:100016. doi: 10.1016/j.bbadva.2021.100016. Epub 2021 Jun 20 - Kagiwada, H; Kiboku, T; Matsuo, H; Kitazawa, M; Fukui, K; Horimoto, K.
Assessing the activation/inhibition of tyrosine kinase-related pathways with a newly developed platform.
PROTEOMICS. 2021 Jun 21:e2000251. doi: 10.1002/pmic.202000251 - Moritsugu, K; Takeuchi, K; Kamiya, N; Higo, J; Yasumatsu, I; Fukunishi, Y; Fukuda, I.
Flexibility and Cell Permeability of Cyclic Ras-Inhibitor Peptides Revealed by the Coupled Nosé-Hoover Equation.
J CHEM INF MODEL. 2021 Apr 9. doi: 10.1021/acs.jcim.0c01427 - Lintuluoto, M; Horioka, Y; Hongo, S; Lintuluoto, JM; Fukunishi, Y.
Molecular Dynamics Simulation Study on Allosteric Regulation of CD44-Hyaluronan Binding as a Force Sensing Mechanism.
ACS OMEGA. 2021 Mar 16;6(12):8045-8055. doi: 10.1021/acsomega.0c05502. eCollection