Biosynthetic Systems Diversity Research Group
Biosynthetic Systems Diversity Research Group Overview
Our group's mission is to use the potential of organisms and our knowledge of chemistry to solve problems in society. We aim to contribute to the development of academia by elucidating biological systems in microorganisms, plants, and animals, and to develop biological products and technologies that contribute to the bio-process and drug discovery based on this understanding, thus contributing to the achievement of a bioeconomical society.

Research Project
Project 1: Drug screening with World-largest natural library
Researcher: KUDO Kei , SAKAMOTO Ryota
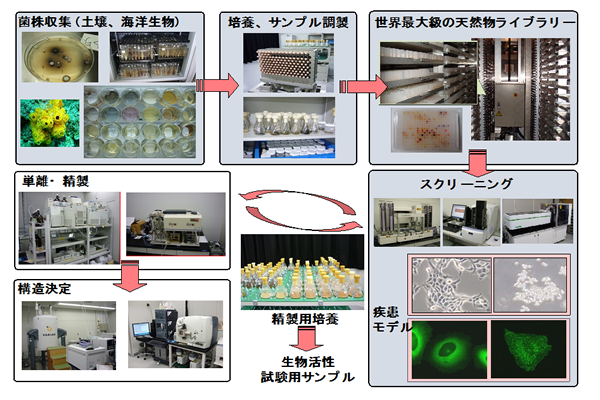
: Natural product library has been applied for drug development, therefore more than 60 percent of the clinical drugs are originated from natural compounds. Pharmaceutical companies provided their natural product library establishing World-largest natural library in our laboratory. We are developing wide range of drug screening for clinical drugs, animal drugs, agricultural chemicals, etc., on various drug targets provided by not only industrial companies but also basic research academia.
Project 2: Development of heterologous expression system for the biosynthesis of natural compounds
Researcher: KUDO Kei, SRISAWAT Pisanee, SAKAMOTO Ryota
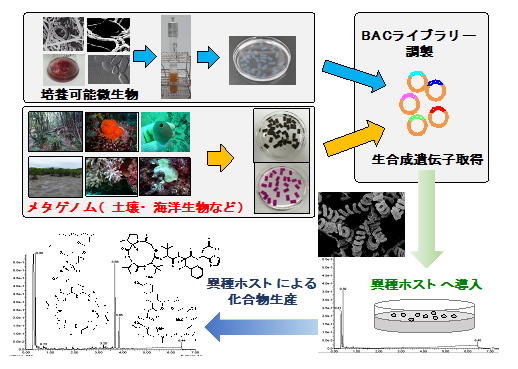
Recent studies revealed that human beings can draw only 1/3 of total ability of the natural compound production by conventional fermentation technology. Our research program targets the development of the next-generation natural compound production technology which leads the production of novel compounds by heterologous expression system applying on cryptic biosynthetic genes. In addition, we perform natural compounds with complex structures which is essential for fine tunings in the drug developments such as improvement of pharmacokinetics, avoidance of side effects and so on. By employing these techniques, we contribute to drug developments.
Project 3:Derivatizing natural products by highly-precise genetic manipulation
Researcher: KUDO Kei, SRISAWAT Pisanee
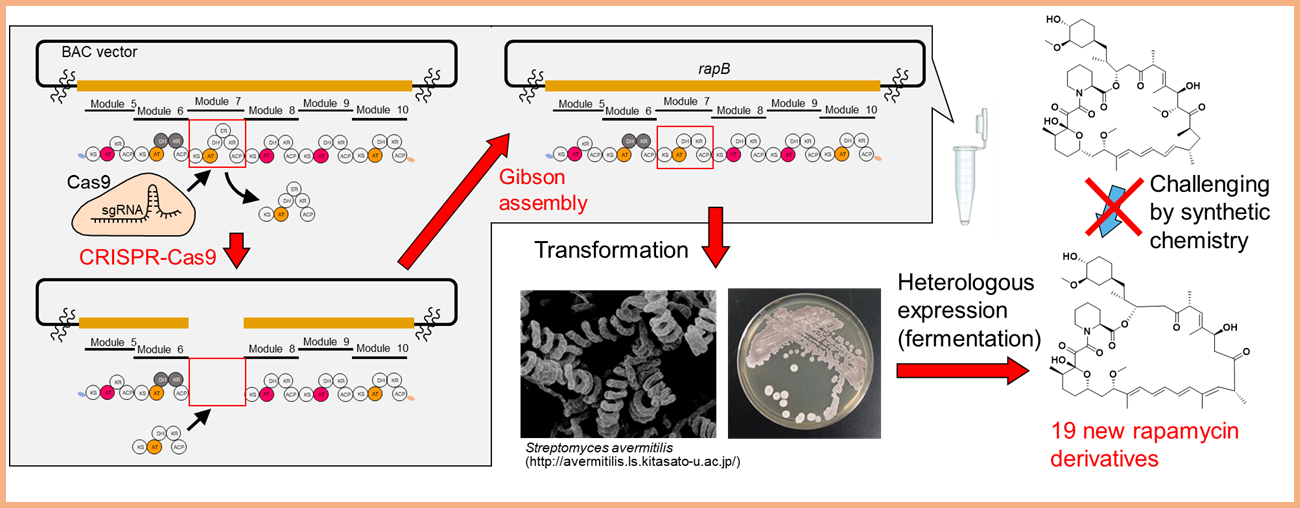
Natural products are an essential resource of bioactive compounds for developing medicines. Such natural products of highly complex structures are challenging to derivatize even by current organic chemistry, so a new methodology is necessary. Recently, we established a genetic engineering technique (in vitro module editing) capable of precisely manipulating the biosynthetic genes for "polyketides." Since the genes encoding polyketide synthase consist of multiple and highly similar sequences, a precise modification was impossible with a conventional recombination-based technique. Our new method makes genetic insertion, replacement, and deletion consistent with the design. It leads to the production of new polyketides.Based on this technology, we are committing to a burst increment of polyketide diversity (combinatorial biosynthesis), fusing natural products with new medical modalities, developing high-valued compounds for stimulating bio-production society, and so on.
Project 4:Elucidation of the function and biosynthetic diversity of modified bases of RNA and its application
Researcher: SHIGI Naoki
The importance of post-translational modifications of proteins and post-transcriptional modifications of RNAs has attracted much attention in recent years in order for genome functions to be performed precisely. We are studying the diversity of RNA modification pathways and their significance in vivo, as many cases of disease with abnormal RNA modifications have been reported in recent years, and we aim to discover new seeds for drug discovery through original basic research. We also aim to create antibiotics without side effects by clarifying the differences in modification mechanisms among species.
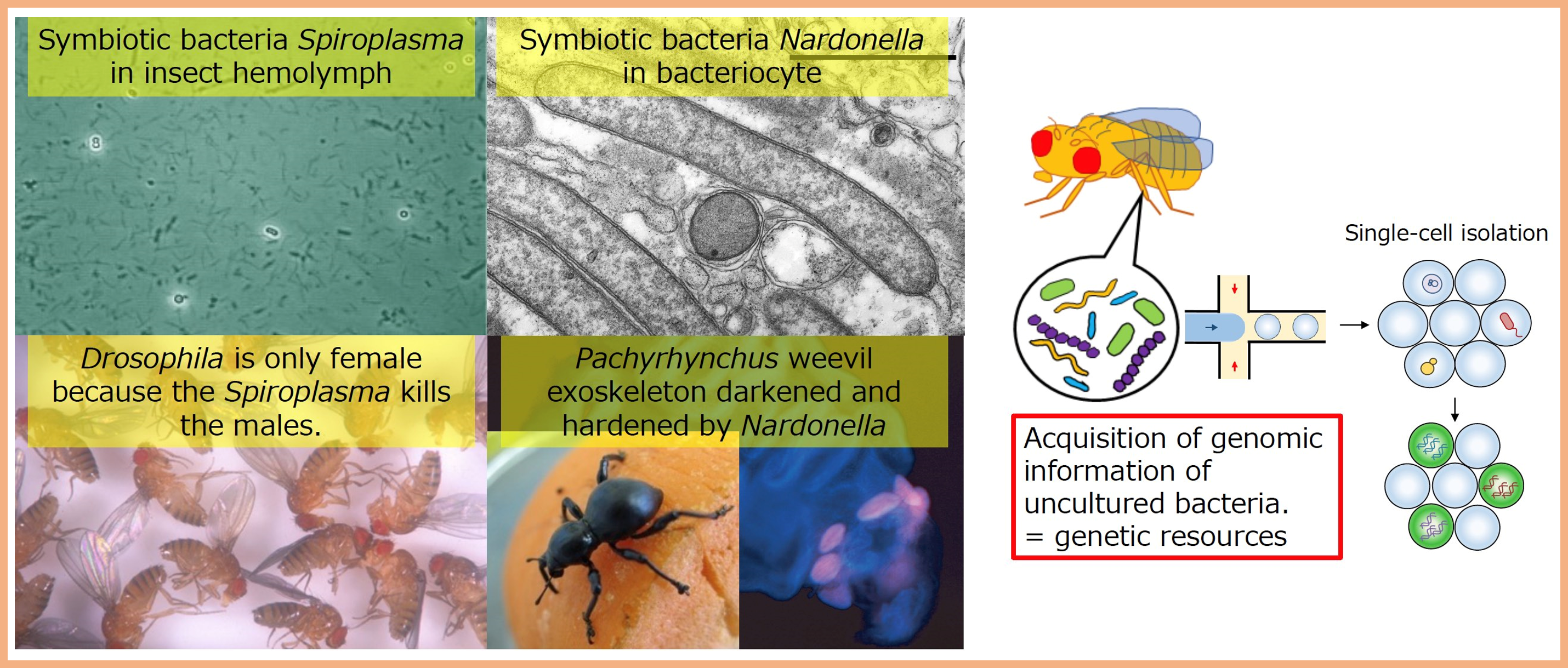
Project 5:Functional analysis of insect symbiotic bacteria and development of a technological basis for their application.
Researcher: ANBUTSU Hisashi
Various symbiotic microorganisms are present in the bodies of many insects and have diverse effects on the physiological functions of their hosts. For example, some symbionts are essential for the hardening of the insect's exoskeleton, while others alter the reproductive mode of the host insect. We mainly work on elucidating their molecular mechanisms through single-cell genome analysis and experiments using model symbiosis systems. Insects are closely related to human society as pests and beneficial insects, and Drosophila and other insects are used as disease model organisms. Therefore, it is hoped that this will lead to the development of fundamental technologies that contribute to the control and pest management of insect populations and support drug discovery and therapy.
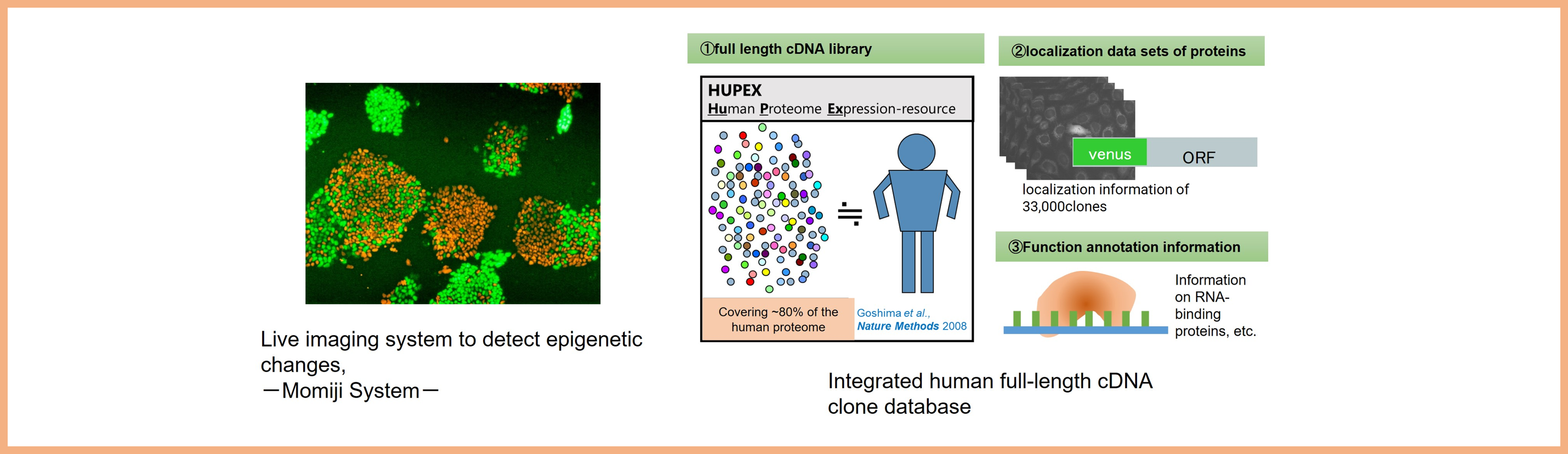
Project 6:Analysis platform for epigenetics and proteomics for understanding cell state
Researcher: KOBAYASHI Shin
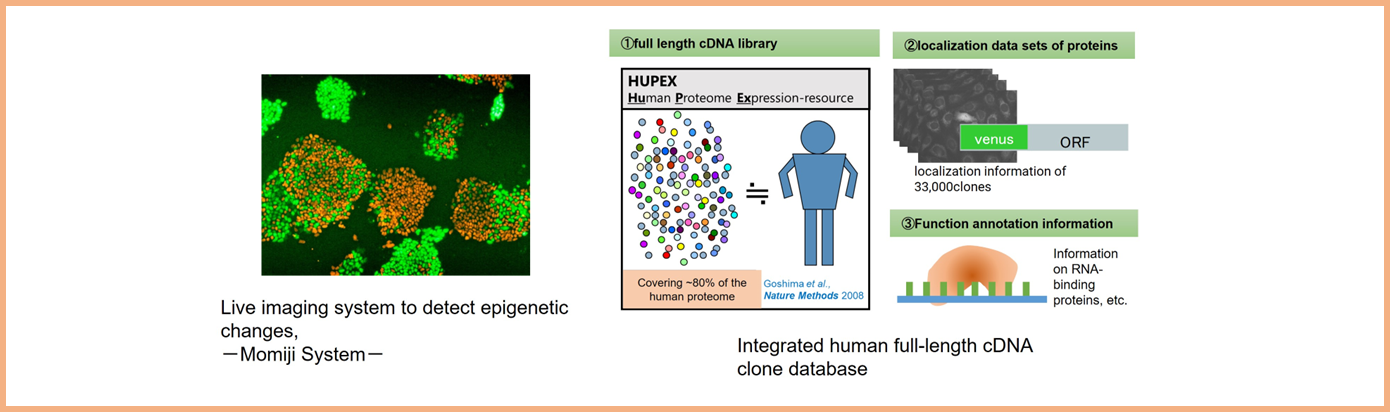
Dynamic cellular events beyond genetic encodings, such as epigenetics, and the formation of membraneless organelles, play critical roles in dictating cellular functions. Taking advantage of technologies such as i) live imaging of epigenetic state, using the Momiji system, and ii) Integrated human full-length cDNA clone database that covers 80% of the human proteome and contains their localization information, we study the dynamics of cell state and promote research that will lead to the understanding of organisms and drug discovery. Specifically, our research focuses on "X chromosome inactivation" in mammals and "non-membrane structures" in the nucleus of mammals.
Member
| photo | position & name | field of expertise | and other info |
|---|---|---|---|
 |
Research Group Leader SHIGI Naoki |
|
|
 |
Senior Researcher ANBUTSU Hisashi |
|
|
 |
Senior Researcher KOBAYASHI Shin |
|
|
 |
Senior Researcher KUDO Kei |
|
|
 |
Researcher SRISAWAT Pisanee |
|
|
 |
Researcher SAKAMOTO Ryota |
|
Results
- Mori, T; Zhou, MY; Kunugitani, K; Akatsuka, T; Yoshida, Y; Kouyama-Suzuki, E; Kobayashi, S; Shirai, Y; Tabuchi, K.
The Competitive Loss of Cerebellar Granule and Purkinje Cells Driven by X-Linked Mosaicism in a Female Mouse Model of CASK-Related Disorders.
CELLS. 2025 May 17;14(10):735. doi:10.3390/cells14100735 - Koshiguchi, M; Yonezawa, N; Hatano, Y; Suenaga, H; Yamagata, K; Kobayashi, S.
A system to analyze the initiation of random X-chromosome inactivation using time-lapse imaging of single cells.
SCI REP. 2024 Sep 2;14(1):20327. doi: 10.1038/s41598-024-71105-y - Nishimura, T; Kudo, K; Izumikawa, M; Kozone, I; Hashimoto, J; Kagaya, N; Suenaga, H; Takeuchi, K; Shin-Ya, K.
Isolation and Structure Elucidation of JBIR-157, a Skeletally Novel Aromatic Polyketide Produced by the Heterologous Expression of a Cryptic Gene Cluster.
CHEM PHARM BULL (Tokyo). 2024;72(5):475-479. doi: 10.1248/cpb.c24-00144 - Kudo, K; Nishimura, T; Izumikawa, M; Kozone, I; Hashimoto, J; Fujie, M; Suenaga, H; Ikeda, H; Satoh, N; Shin-Ya, K.
Capability of a large bacterial artificial chromosome clone harboring multiple biosynthetic gene clusters for the production of diverse compounds.
J ANTIBIOT (Tokyo). 2024 Mar 4. doi: 10.1038/s41429-024-00711-9 - Sugio, Y; Yamagami, R; Shigi, N; Hori, H.
A selective and sensitive detection system for 4-thiouridine modification in RNA.
RNA. 2022 Nov 21:rna.079445.122. doi: 10.1261/rna.079445.122 - Choirunnisa, AR; Arima, K; Abe,Y; Kagaya, K; Kudo, K; Suenaga, H; Hashimoto, J; Fujie, M; Satoh, N; Shin-Ya, K; Matsuda, K; Wakimoto, T.
New azodyrecins identified by a genome mining-directed reactivity-based screening.
BEILSTEIN J ORG CHEM. 2022 Aug 10;18:1017-1025. doi: 10.3762/bjoc.18.102 - Kojima, S; Shiochi, N; Sato, K; Yamaura, M; Ito, T; Yamamura, N; Goto, N; Odamoto, M; Kobayashi, S; Kimura, T; Sekita, Y.
Epigenome editing reveals core DNA methylation for imprinting control in the Dlk1-Dio3 imprinted domain.
NUCLEIC ACIDS RESEARCH. 2022 May 11. doi: 10.1093/nar/gkac344 - Aizawa, S; Nishimura, K; Mondejar, GS; Kumar, A; Bui, PL; Tran, YTH; Kuno, A; Muratani, M; Kobayashi, S; Nabekura, T; Shibuya, A; Sugihara, E; Sato, TA; Fukuda, A; Hayashi, Y; Hisatake, K.
Early reactivation of clustered genes on the inactive X chromosome during somatic cell reprogramming.
STEM CELL REPORTS. 2021 Nov 29:S2213-6711(21)00589-0. doi10.1016/j.stemcr.2021.11.008 - Zhang, M; Kiyono, T; Aoki, K; Goshima, N; Kobayashi, S; Hiranuma, K; Shiraishi, K; Saya, H; Nakahara, T.
Development of an in vitro carcinogenesis model of HPV induced cervical adenocarcinoma.
CANCER SCI. 2021 Dec 21. doi: 10.1111/cas.15246 - Shigi, N.
Biosynthesis and Degradation of Sulfur Modifications in tRNAs.
INT J MOL SCI. 2021 Nov 3;22(21):11937. doi: 10.3390/ijms222111937 - Kudo, K; Nishimura, T; Kozone, I; Hashimoto, J; Kagaya, N; Suenaga, H; Ikeda, H; Shin-Ya, K.
Hemiacetal-less rapamycin derivatives designed and produced by genetic engineering of a type I polyketide synthase.
SCI REP. 2021 May 11;11(1):9944. doi: 10.1038/s41598-021-88583-z - Liu, C; Hashimoto, J; Kudo, K; Shin-Ya, K; Kakeya, H.
An Atypical Arginine Dihydrolase Involved in the Biosynthesis of Cyclic Hexapeptide Longicatenamides.
CHEM ASIAN J. 2021 Apr 22. doi: 10.1002/asia.202100181 - Haramoto, Y; Sakata, M; Kobayashi, S.
Visualization of X chromosome reactivation in mouse primordial germ cells in vivo.
BIOL OPEN. 2021 Apr 15;10(4):bio058602. doi: 10.1242/bio.058602. Epub 2021 Apr 29