Cell Dynamics System Research Group
Cell Dynamics System Research Group Overview
In the cell industry, including bio-pharmaceuticals, regenerative medicine, and cell-based foods, new cell control technologies are required, distinct from traditional manufacturing processes. To optimize product development and manufacturing processes in the cell industry, we aim to integrate the cell/animal dynamics analysis and omics data analysis, and develop new technologies necessary for cell manipulation, modification, and quality control.
Research Project
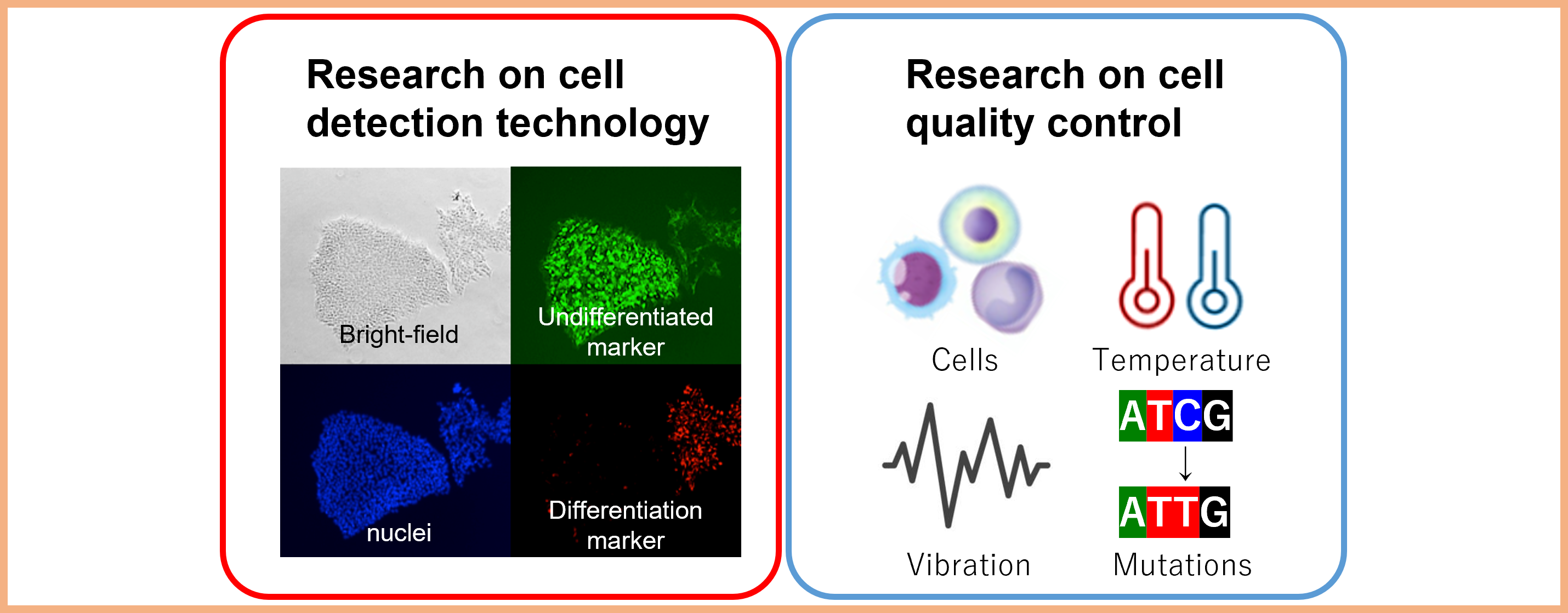
Project 1:Development of technology for the manipulation and management of industrial cells
Researcher: MAWARIBUCHI Shuuji
We are developing cell manipulation and management technologies for manufacturing processes in industries that use cells, such as regenerative medicine and cellular food.
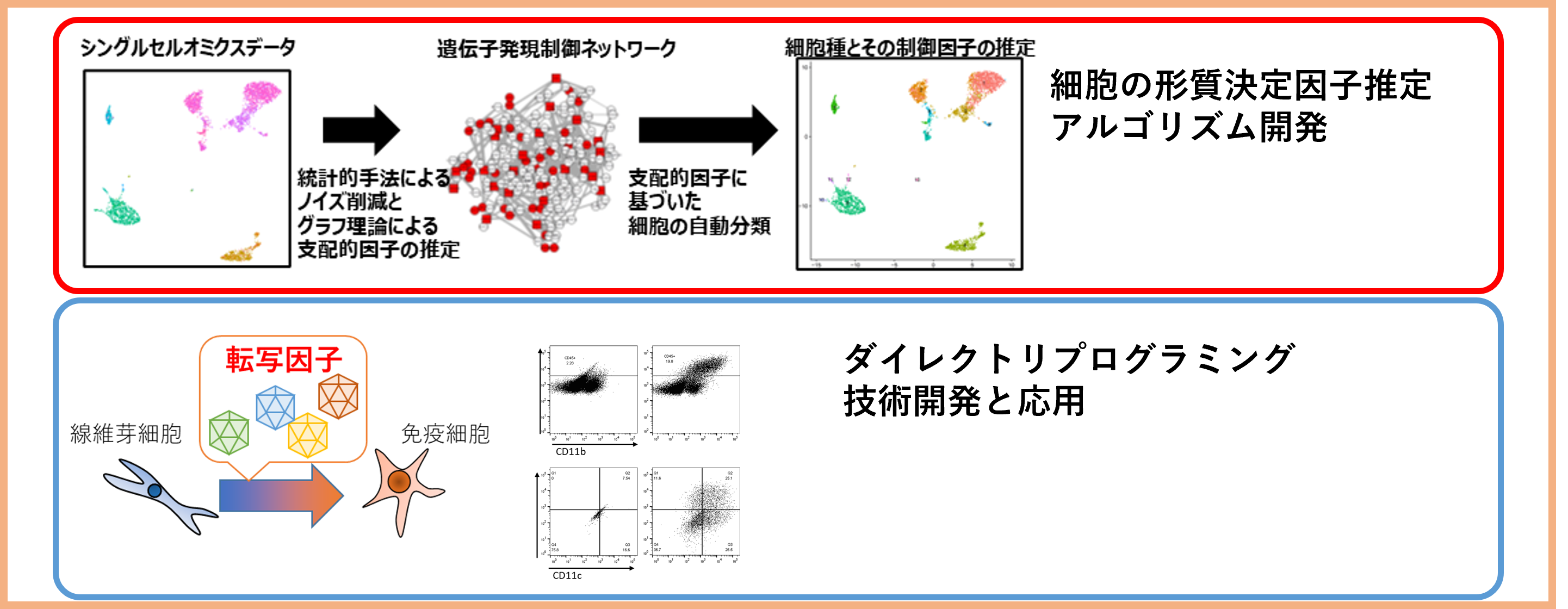
Project 2:Development and application of algorithm inferring cell phenotype determining factors
Researcher: KUMAGAI Yutaro
We are developing algorithm to infer factors determining cell phenotype from multimodal omics data. We apply the algorithms to control cell phenotype.
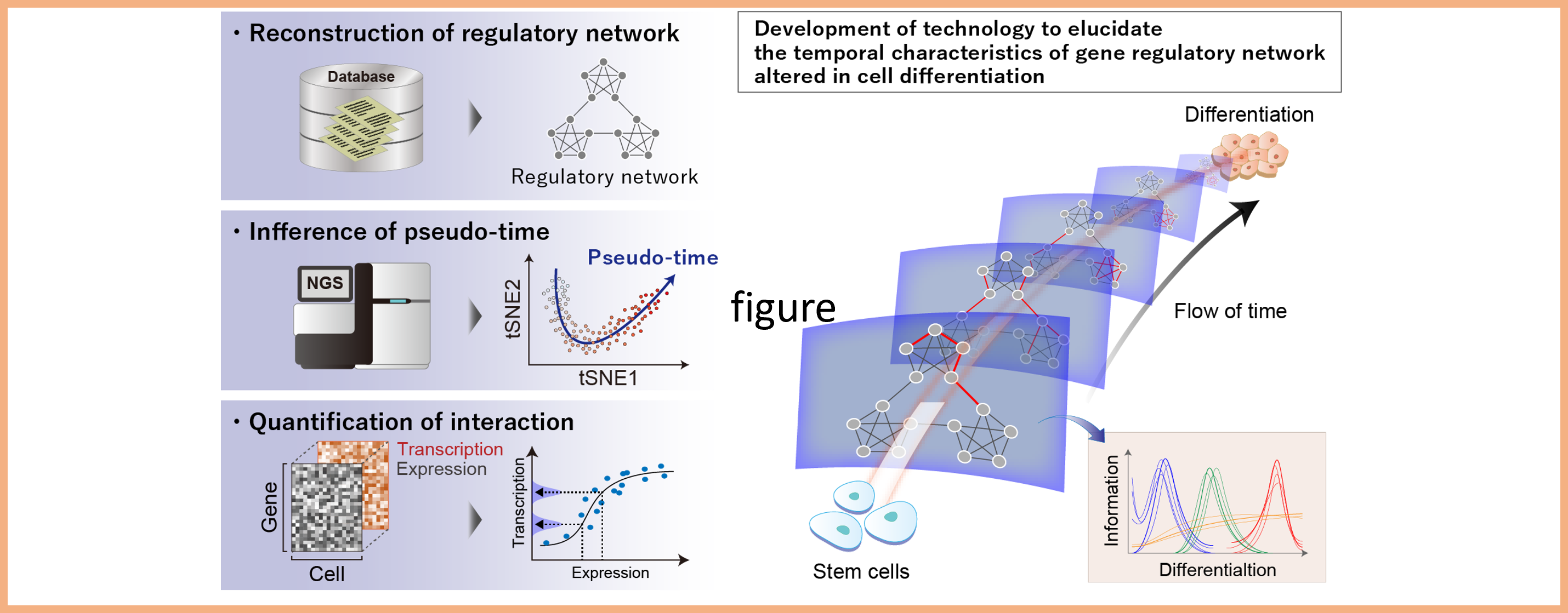
Project 3:Development of technology to analyze dynamic properties in cellular regulatory networks
Researcher: KAWATA Kentaro
Various kinds of molecules consist of cells, and they transmit information to determine the cellular state. In this research, we are developing technologies to elucidate precisely how the molecular network alters accompanied by cell differentiation and adaptation to the extracellular environment, and how the changes of the network affect the cellular state. Through these studies, we promote highly accurate regulation of differentiation in pluripotent stem cells.
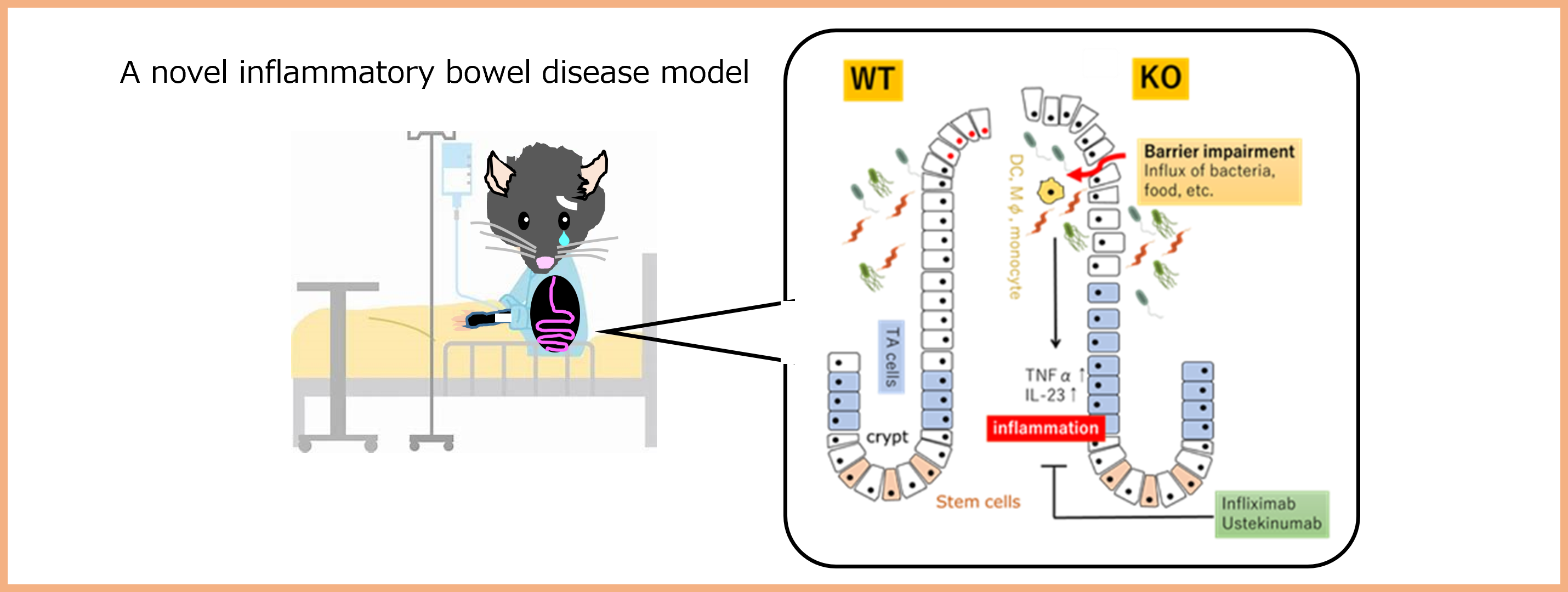
Project 4:Making a “society of health and longevity” a reality: etiology of chronic inflammatory diseases
Researcher: SASAKI Yasunori
We have been working on a protein that involves in a battery of biological processes and diseases. Mice lack this protein suffer impairment of the intestinal barrier that leads to many chronic inflammatory diseases. This mouse would be a versatile tool to investigate various diseases caused by chronic inflammation, e. g. inflammatory bowel disease.
Staff Members
| photo | position & name | field of expertise | and other info |
|---|---|---|---|
 |
Research Group Leader MAWARIBUCHI Shuuji |
|
|
 |
Senior Researcher SASAKI Yasunori |
|
|
 |
Senior Researcher KUMAGAI Yutaro |
|
|
 |
Attached to Research Group KAWATA Kentaro |
|
Results
- Zhang, Y; Yang, W; Kumagai, Y; Loza, M; Yang, Y; Park, SJ; Nakai, K.
In Silico Analysis Revealed Marco (SR-A6) and Abca1/2 as Potential Regulators of Lipid Metabolism in M1 Macrophage Hysteresis.
INT J MOL SCI. 2024 Dec 26. doi:10.3390/ijms26010111 - Kusano, T; Sotani, Y; Takeda, R; Hatano, A; Kawata, K; Kano, R; Matsumoto, M; Kano, Y; Hoshino, D.
Time-series transcriptomics reveals distinctive mRNA expression dynamics associated with gene ontology specificity and protein expression in skeletal muscle after electrical stimulation-induced resistance exercise.
FASEB J. 2024 Nov 30;38(22):e70153. doi:10.1096/fj.202401420RR - Zhou, J; Sekiguchi, Y; Sano, M; Nishimura, K; Hisatake, K; Fukuda, A.
A Sendai virus-based expression system directs efficient induction of chondrocytes by transcription factor-mediated reprogramming.
SCI REP. 2024 Oct 29;14(1):26004. doi: 10.1038/s41598-024-77508-1 - Mawaribuchi, S; Iida, M; Haramoto, Y.
Fusion of breast cancer MCF-7 cells with mesenchymal stem cells rearranges interallelic gene expression and enhances cancer malignancy.
BIOCHEM BIOPHYS RES COMMUN. 2024 Oct 23:736:150887. doi: 10.1016/j.bbrc.2024.150887 - Kumagai, Y.
BootCellNet, a resampling-based procedure, promotes unsupervised identification of cell populations via robust inference of gene regulatory networks.
PLOS COMPUT BIOL. 2024 Sep 30;20(9):e1012480. doi: 10.1371/journal.pcbi.1012480 - Miura, I; Shams, F; Ohki, J; Tagami, M; Fujita, H; Kuwana, C; Nanba, C; Matsuo, T Ogata, M; Mawaribuchi, S; Shimizu, N; Ezaz, T.
Multiple Transitions between Y Chromosome and Autosome in Tagos Brown Frog Species Complex.
GENES (Basel). 2024 Feb 26;15(3):300. doi: 10.3390/genes15030300 - Kishimoto, T; Nishimura, K; Morishita, K; Fukuda, A; Miyamae, Y; Kumagai, Y; Sumaru, K; Nakanishi, M; Hisatake, K; Sano, M.
An engineered ligand-responsive Csy4 endoribonuclease controls transgene expression from Sendai virus vectors.
J BIOL ENG. 2024 Jan 16;18(1):9. doi: 10.1186/s13036-024-00404-9 - Zhang, Y; Yang, W; Kumagai, Y; Loza, M; Zhang, W; Park, SJ; Nakai, K.
Multi-omics computational analysis unveils the involvement of AP-1 and CTCF in hysteresis of chromatin states during macrophage polarization.
FRONT IMMUNOL. 2023 Dec 20;14:1304778. doi: 10.3389/fimmu.2023.1304778 - Takeda, N; Tsuchiya, A; Mito, M; Natsui, K; Natusi, Y; Koseki, Y; Tomiyoshi, K; Yamazaki, F; Yoshida, Y; Abe, H; Sano, M; Kido, T; Yoshioka, Y; Kikuta, J; Itoh, T; Nishimura, K; Ishii, M; Ochiya, T; Miyajima, A; Terai, S.
Analysis of distribution, collection, and confirmation of capacity dependency of small extracellular vesicles toward a therapy for liver cirrhosis.
INFLAMM REGEN. 2023 Oct 9;43(1):48. doi: 10.1186/s41232-023-00299-x - Kumagai, Y; Saito, Y; Kida, YS.
A multiomics atlas of brown adipose tissue development over time.
ENDOCRINOLOGY. 2023 Apr 21:bqad064. doi: 10.1210/endocr/bqad064 - Mawaribuchi, S; Ito, M; Ogata, M; Yoshimura, Y; Miura, I.
Parallel Evolution of Sex-Linked Genes across XX/XY and ZZ/ZW Sex Chromosome Systems in the Frog Glandirana rugosa.
GENES (Basel). 2023 Jan 18;14(2):257. doi: 10.3390/genes14020257 - Mawaribuchi, S; Shimomura, O; Oda, T; Hiemori, K; Shimizu, K; Yamase, K; Date, M; Tateno, H.
rBC2LCN-reactive SERPINA3 is a glycobiomarker candidate for pancreatic ductal adenocarcinoma.
GLYCOBIOLOGY. 2023 Feb 2:cwad009. doi: 10.1093/glycob/cwad009 - Huang, T; Sato, Y; Kuramochi, A; Ohba, Y; Sano, M; Miyagishi, M; Tateno, H; Wadhwa, R; Kawasaki, K; Uchida, T; Ekdahl, KN; Nilsson, B; Chung, UI; Teramura, Y.
Surface modulation of extracellular vesicles with cell-penetrating peptide-conjugated lipids for improvement of intracellular delivery to endothelial cells.
REGEN THER. 2023 Jan 11;22:90-98. doi: 10.1016/j.reth.2022.12.007 - Mawaribuchi, S; Haramoto, Y; Ikeda, N; Ito, M.
Evolutionary features of ligands and their receptors via protein-protein interactions and essentiality in primates.
GENES CELLS. 2023 Jan 9. doi: 10.1111/gtc.13006 - Imai, Y;Mori, N; Nihashi, Y; Kumagai, Y; Shibuya, Y; Oshima, J; Sasaki, M; Sasaki, K; Aihara, Y; Sekido, M; Kida, YS.
Therapeutic Potential of Adipose Stem Cell-Derived Conditioned Medium on Scar Contraction Model.
BIOMEDICINES. 2022 Sep 24;10(10):2388. doi: 10.3390/biomedicines10102388 - Sano, M; Morishita, K; Onizawa, Y; Takagi, T; Sumaru, K.
Rapid and Highly Sensitive Method for Evaluating Surface Coatings against an Enveloped RNA Virus.
ACS APPL BIO MATER. 2022 Oct 14. doi: 10.1021/acsabm.2c00613 - Katsumi, T; Shams, F; Yanagi, H; Ohnishi, T; Toda, M; Lin, SM; Mawaribuchi, S; Shimizu, N; Ezaz, T; Miura, I.
Highly rapid and diverse sex chromosome evolution in the Odorrana frog species complex.
DEV GROWTH DIFFER. 2022 Jul 26. doi: 10.1111/dgd.12800 - Omoto, T; Yimiti, D; Sanada, Y; Toriyama, M; Ding, C; Hayashi, Y; Ikuta, Y; Nakasa, T; Ishikawa, M; Sano, M; Lee, M; Akimoto, T; Shukunami, C; Miyaki, S; Adachi, N.
Tendon-Specific Dicer Deficient Mice Exhibit Hypoplastic Tendon Through the Downregulation of Tendon-Related Genes and MicroRNAs.
FRONT CELL DEV BIOL. 2022 Jun 14;10:898428. doi: 10.3389/fcell.2022.898428 - Miura, I; Shams, F; Jeffries, DL; Katsura, Y; Mawaribuchi, S; Perrin, N; Ito, M; Ogata, M; Ezaz, T.
Identification of ancestral sex chromosomes in the frog Glandirana rugosa bearing XX-XY and ZZ-ZW sex-determining systems.
MOL ECOL. 2022 Jun 12. doi: 10.1111/mec.16551 - Sugisawa, E; Kondo, T; Kumagai, Y; Kato, H; Takayama, Y; Isohashi, K; Shimosegawa, E; Takemura, N; Hayashi, Y; Sasaki, T; Martino, MM; Tominaga, M; Maruyama, K.
Nociceptor-derived Reg3γ prevents endotoxic death by targeting kynurenine pathway in microglia.
CELL REP. 2022 Mar 8;38(10):110462. doi: 10.1016/j.celrep.2022.110462 - Sano, M; Morishita, K; Oikawa, S; Akimoto, T; Sumaru, K; Kato, Y.
Live-cell imaging of microRNA expression with post-transcriptional feedback control.
MOL THER NUCLEIC ACIDS. 2021 Aug 26;26:547-556. doi: 10.1016/j.omtn.2021.08.018 - Ogita, Y; Tamura, K; Mawaribuchi, S; Takamatsu, N; Ito, M.
Independent pseudogenizations and losses of sox15 during amniote diversification following asymmetric ohnolog evolution.
BMC ECOL EVOL. 2021 Jun 30;21(1):134. doi: 10.1186/s12862-021-01864-z