Cell Function Design Research Group
Cell Function Design Research Group Overview
Misson of our Group is to develop highly designed and optimized cells that maximize the substance production capacity of biological cells by precisely controlling and modifying their properties. Our group aims to construct cells that are useful for substance production and drug discovery contributing to the creation of a new cell industry.
Research Project
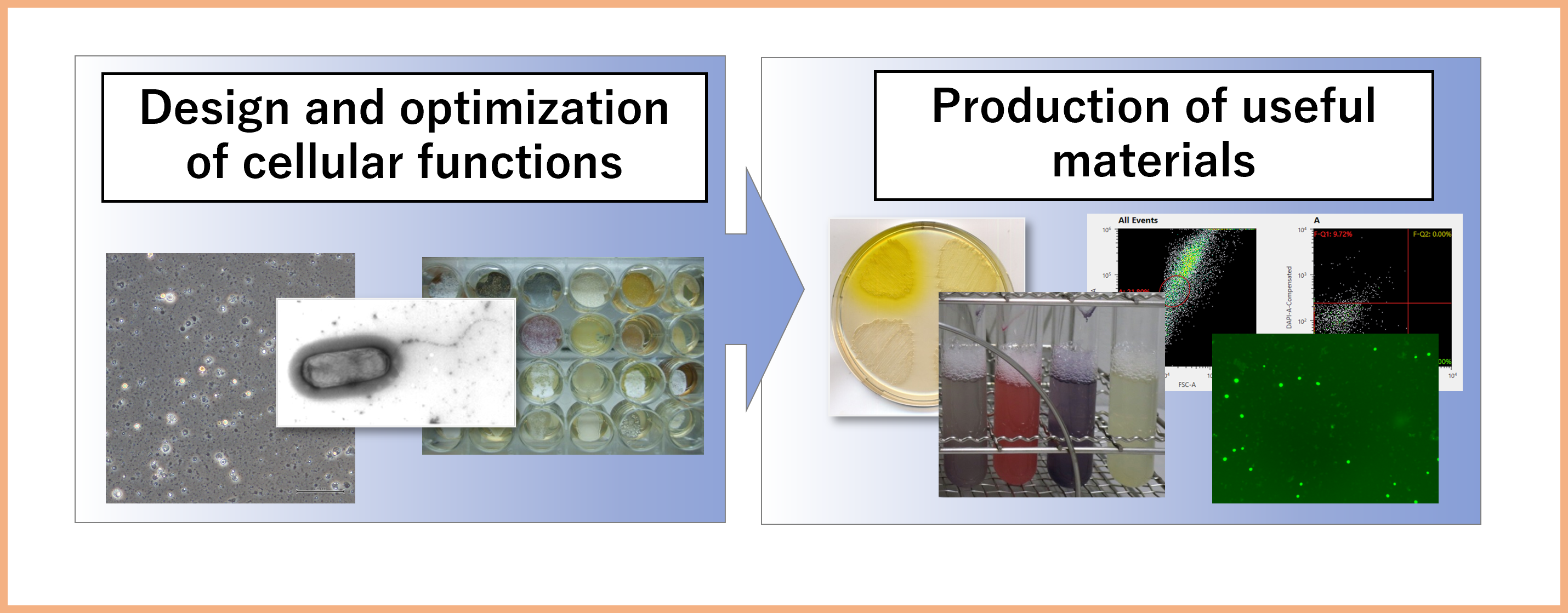
Project 1:Production of useful materials using cells
Researcher: SUENAGA Hikaru
We produce optimized microorganisms and animal cells for the material production through sophisticated design and precise control and modification of cell properties.
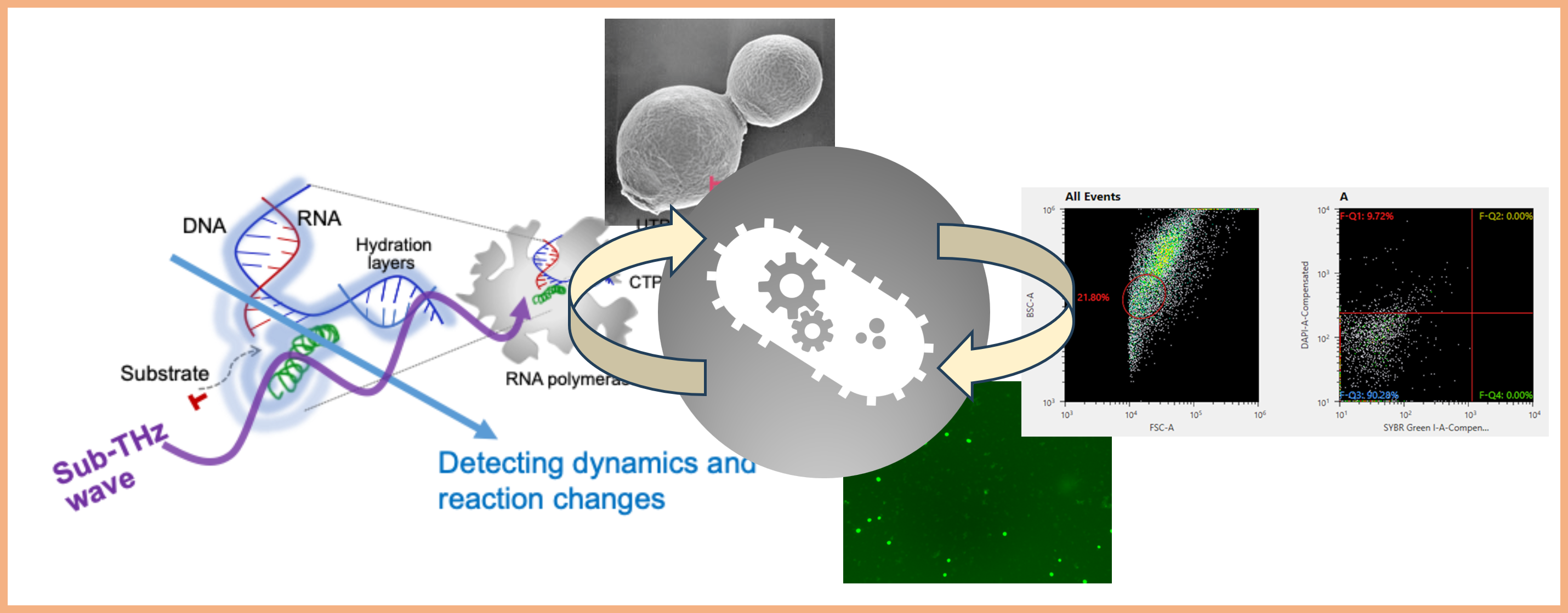
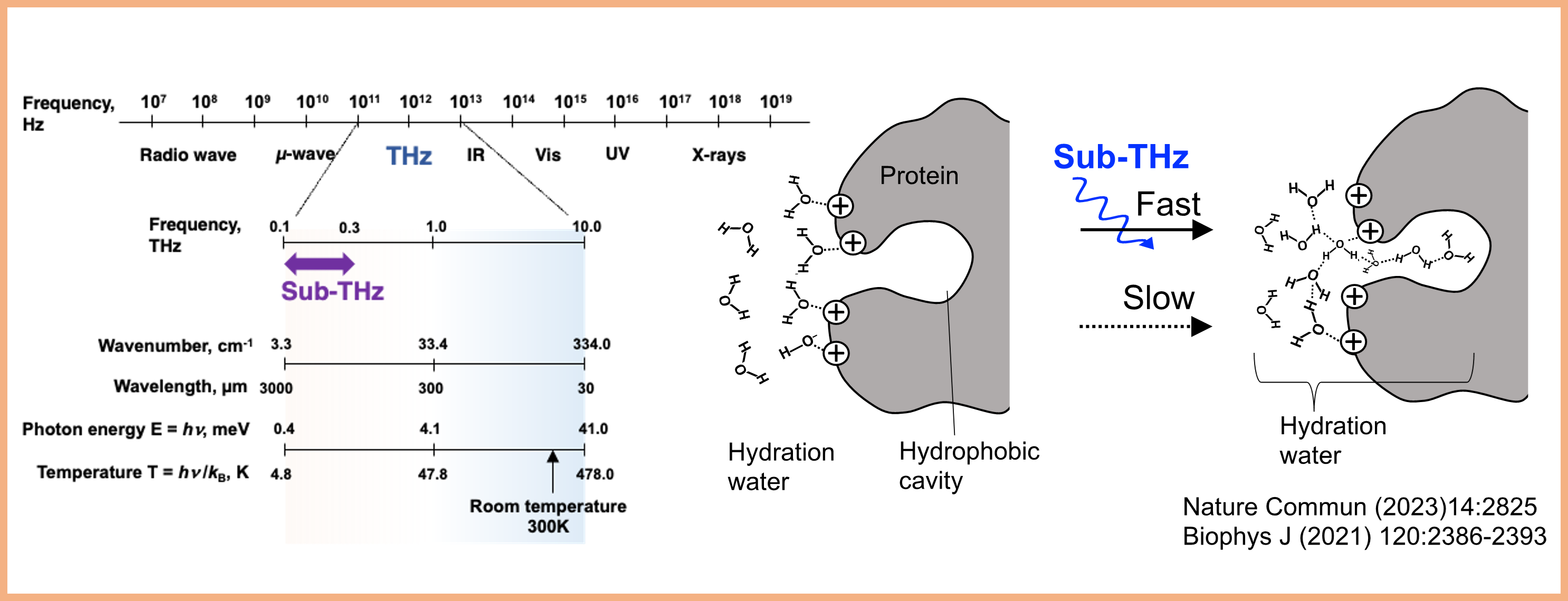
Project 2:Control of molecular and cellular functions and hydration using Sub-THz waves
Researcher: IMASHIMIZU Masahiko
Many of the sophisticated biomolecular functions are produced by the interactions between water and biomacromolecules (mostly hydrogen bonds) and their biased fluctuations. Fluctuations in hydrogen bonds appear in the THz frequency region. From the viewpoint of “control by Sub-THz waves,” we are working to elucidate biomolecular functions and to develop applications that extend to multiple fields.
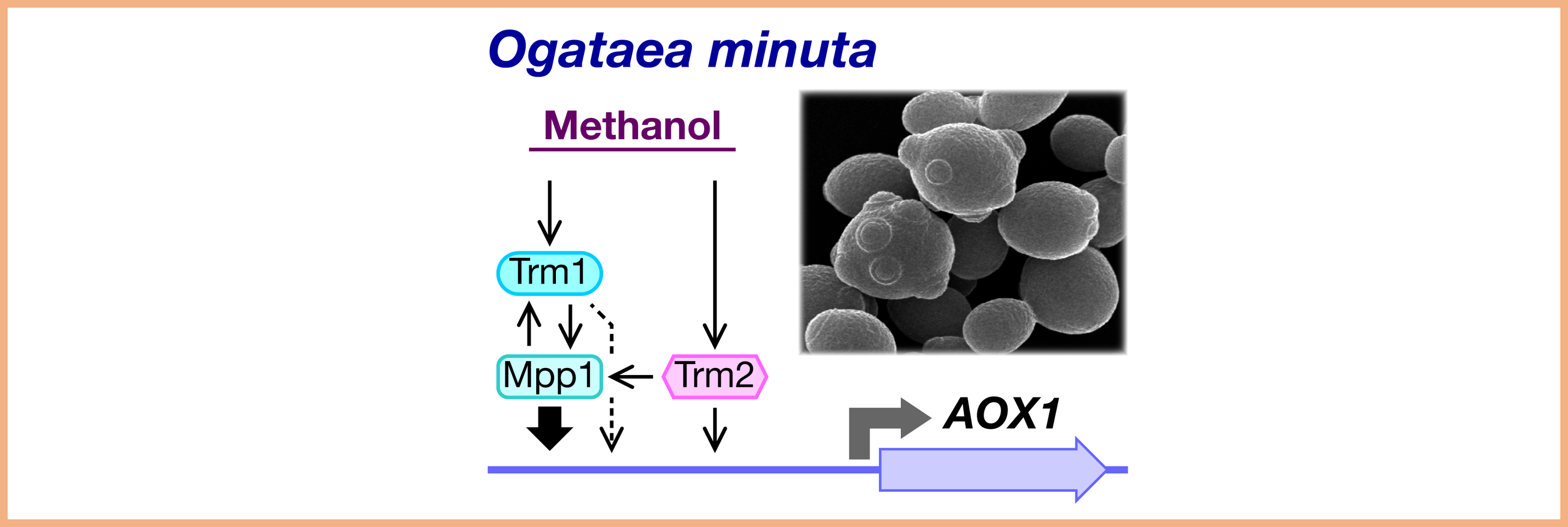
Project 3:Heterologous protein production in yeasts
Researcher: YOKO-O Takehiko
The methanol-utilizing yeast Ogataea minuta is an excellent host for heterologous protein production. Our research focuses on the development of plasmids and libraries, as well as the generation of high-yield protein-producing strains in this species. Additionally, we study the biosynthetic mechanisms of glycosylphosphatidylinositol (GPI) anchors and the localization mechanisms of glycosyltransferases.
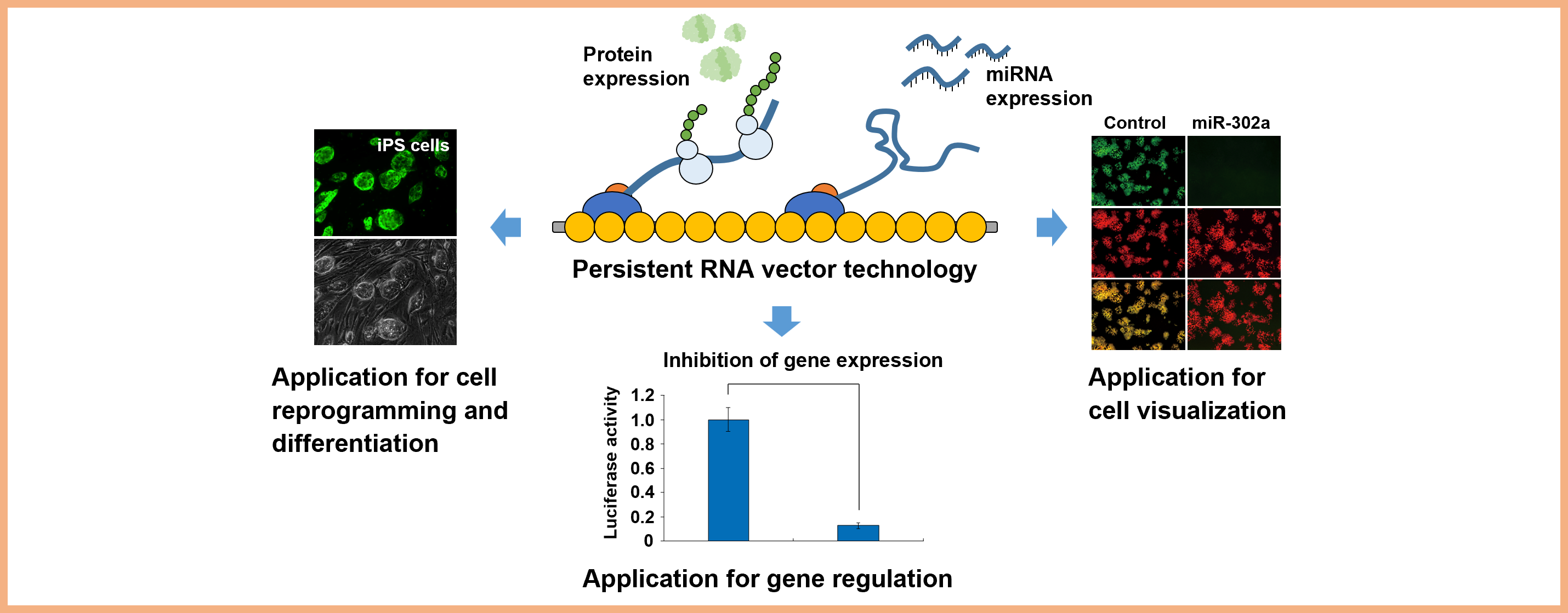
Project 4:Development of an RNA vector for efficient gene delivery and biological applications
Researcher: SANO Masayuki
We have been developing the efficient gene delivery system that leads to high transduction efficiency, long-term expression of coding- and non-coding genes, and control of transgene expression using the persistent RNA vector. The vector system can be used to facilitate cell visualization, regulation of endogenous gene expression, and cell reprogramming and differentiation.
Staff Members
| photo | position & name | field of expertise | and other info |
|---|---|---|---|
 |
Research Group Leader SUENAGA Hikaru |
|
|
 |
Senior Researcher TAKAHASHI Katsutoshi |
|
|
 |
Senior Researcher SANO Masayuki |
|
|
 |
Chief Senior Researcher IMASHIMIZU Masahiko |
|
|
 |
Attached to Research Group YOKO-O Takehiko |
|
Results
- Zhu, J; Cai, HL; Goto, S; Shimada, M; Yurimoto, H; Sakai, Y; Yoko-O, T; Chiba, Y; Nakagawa, T.
Physiological role of the FDH1 gene in methylotrophic yeast Komagataella phaffii and carbon recovery as formate during methylotrophic growth of the fdh1-deletion strain.
J BIOSCI BIOENG. 2025 Jan 17. doi:10.1016/j.jbiosc.2024.12.007 - Hashimoto, T; Suenaga, H; Shin-Ya, K.
Application of Cas9-Based Gene Editing to Engineering of Nonribosomal Peptide Synthetases.
CHEMBIOCHEM. 2024 Dec 30:e202400765. doi:10.1002/cbic.202400765 - Kudo, K; Nishimura, T; Miyako, K; Suenaga, H; Shin-Ya, K.
A new cannabigerolic acid derivative and its unprenylated precursor produced through the reconstitution of cannabinoid biosynthesis in Streptomyces.
J ANTIBIOT (Tokyo). 2024 Dec 3. doi:10.1038/s41429-024-00793-5 - Yamamoto, K; Uzaki, M; Takahashi, K; Mimura, T.
Current status of MSI research in Japan to measure the localization of natural products in plants.
CURR OPIN PLANT BIOL. 2024 Oct 19;82:102651. doi: 10.1016/j.pbi.2024.102651 - Koshiguchi, M; Yonezawa, N; Hatano, Y; Suenaga, H; Yamagata, K; Kobayashi, S.
A system to analyze the initiation of random X-chromosome inactivation using time-lapse imaging of single cells.
SCI REP. 2024 Sep 2;14(1):20327. doi: 10.1038/s41598-024-71105-y - Song, X; Nihashi, Y; Imai, Y; Mori, N; Kagaya, N; Suenaga, H; Shin-Ya, K; Yamamoto, M; Setoyama, D; Kunisaki Y; Kida, YS .
Collagen Lattice Model, Populated with Heterogeneous Cancer-Associated Fibroblasts, Facilitates Advanced Reconstruction of Pancreatic Cancer Microenvironment.
INT J MOL SCI. 2024 Mar 27;25(7):3740. doi: 10.3390/ijms25073740 - Kudo, K; Nishimura, T; Izumikawa, M; Kozone, I; Hashimoto, J; Fujie, M; Suenaga, H; Ikeda, H; Satoh, N; Shin-Ya, K.
Capability of a large bacterial artificial chromosome clone harboring multiple biosynthetic gene clusters for the production of diverse compounds.
J ANTIBIOT (Tokyo). 2024 Mar 4. doi: 10.1038/s41429-024-00711-9 - Qayyum, MZ; Imashimizu, M; Leanca, M; Vishwakarma, RK; Riaz-Bradley, A; Yuzenkova, Y; Murakami, KS.
Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
PROC NATL ACAD SCI U S A. 2024 Feb 20;121(8):e2311480121. doi: 10.1073/pnas.2311480121 - Hashimoto, T; Suenaga, H; Amagai, K; Hashimoto, J; Kozone, I; Takahashi, S; Shin-Ya, K.
In Vitro Module Editing Of NRPS Enables Production Of Highly Potent Gq-Signaling Inhibitor FR900359 Derived From Unculturable Plant Symbiont.
ANGEW CHEM INT ED ENGL. 2024 Jan 18:e202317805. doi: 10.1002/anie.202317805 - Suzuki, S; Saito, S; Narushima, Y; Kodani, S; Kagaya, N; Suenaga, H; Shin-Ya, K; Arai, MA.
Notch activator cyclopiazonic acid induces apoptosis in HL-60 cells through calcineurin activation.
J ANTIBIOT (Tokyo). 2023 Nov 8. doi: 10.1038/s41429-023-00673-4 - Sugiyama, JI; Tokunaga, Y; Hishida, M; Tanaka, M; Takeuchi, K; Satoh, D; Imashimizu, M.
Nonthermal acceleration of protein hydration by sub-terahertz irradiation.
NAT COMMUN. 2023 May 22;14(1):2825. doi: 10.1038/s41467-023-38462-0 - Fujihara, H; Hirose, J; Suenaga, H.
Evolution of genetic architecture and gene regulation in biphenyl/PCB-degrading bacteria.
FRONTIERS IN MICROBIOLOGY. 2023 Jun 7;14:1168246. doi: 10.3389/fmicb.2023.1168246 - Tsuda, M; Nakatani, Y; Baba, S; Tanaka, I; Ichikawa, K; Nonaka, K; Ito, R; Yoko-O, T; Chiba, Y.
Selection of the methylotrophic yeast Ogataea minuta as a high-producing host for heterologous protein expression.
J BIOSCI BIOENG. 2023 Jan 24:S1389-1723(22)00375-9. doi: 10.1016/j.jbiosc.2022.12.006 - Choirunnisa, AR; Arima, K; Abe,Y; Kagaya, K; Kudo, K; Suenaga, H; Hashimoto, J; Fujie, M; Satoh, N; Shin-Ya, K; Matsuda, K; Wakimoto, T.
New azodyrecins identified by a genome mining-directed reactivity-based screening.
BEILSTEIN J ORG CHEM. 2022 Aug 10;18:1017-1025. doi: 10.3762/bjoc.18.102 - Mellul, M; Lahav, S; Imashimizu, M; Tokunaga, Y; Lukatsky, DB; Ram, O.
Repetitive DNA symmetry elements negatively regulate gene expression in embryonic stem cells.
BIOPHYS J. 2022 Jul 8:S0006-3495(22)00557-4. doi: 10.1016/j.bpj.2022.07.011 - Matsuda, K; Arima, K; Akiyama, S; Yamada, Y; Abe, Y; Suenaga, H; Hashimoto, J; Shin-Ya, K; Nishiyama, M; Wakimoto, T.
A Natural Dihydropyridazinone Scaffold Generated from a Unique Substrate for a Hydrazine-Forming Enzyme.
J AM CHEM SOC. 2022 Jun 30. doi: 10.1021/jacs.2c05269 - Yamamoto, K; Takahashi, K; OConnor, SE; Mimura, T.
Imaging MS Analysis in Catharanthus roseus.
METHODS MOL BIOL. 2022;2505:33-43. doi: 10.1007/978-1-0716-2349-7_2 - Suenaga, H; Matsuzawa, T; Sahara, T.
Discovery by metagenomics of a functional tandem repeat sequence that controls gene expression in bacteria.
FEMS MICROBIOL ECOL. 2022 Mar 28:fiac037. doi: 10.1093/femsec/fiac037 - Hirose, J; Watanabe, T; Futagami, T; Fujihara, H; Kimura, N; Suenaga, H; Goto, M; Suyama, A; Furukawa, K.
A New ICEclc Subfamily Integrative and Conjugative Element Responsible for Horizontal Transfer of Biphenyl and Salicylic Acid Catabolic Pathway in the PCB-Degrading Strain Pseudomonas stutzeri KF716.
MICROORGANISMS. 2021 Nov 29;9(12):2462. doi: 10.3390/microorganisms9122462 - Yoko-O, T; Komatsuzaki, A; Yoshihara, E; Zhao, S; Umemura, M; Gao, XD; Chiba, Y.
Regulation of alcohol oxidase gene expression in methylotrophic yeast Ogataea minuta.
J BIOSCI BIOENG. 2021 Aug 27:S1389-1723(21)00204-8. doi: 10.1016/j.jbiosc.2021.08.001 - Suenaga, H; Kagaya, N; Kawada, M; Tatsuda, D; Sato, T; Shin-Ya, K.
Phenotypic screening system using three-dimensional (3D) culture models for natural product screening.
J ANTIBIOT (Tokyo). 2021 Jul 29. doi: 10.1038/s41429-021-00457-8 - Nishimura, T; Kawahara, T; Kagaya, N; Ogura, Y; Takikawa, H; Suenaga, H; Adachi, M; Hirokawa, T; Doi, T; Shin-Ya, K.
JBIR-155, a Specific Class D β-Lactamase Inhibitor of Microbial Origin.
ORG LETT. 2021 May 24. doi: 10.1021/acs.orglett.1c01352 - Tokunaga, Y; Tanaka, M; Iida, H; Kinoshita, M; Tojima, Y; Takeuchi, K; Imashimizu, M.
Nonthermal excitation effects mediated by subterahertz radiation on hydrogen exchange in ubiquitin
BIOPHYS J. 2021 Apr 21:S0006-3495(21)00329-5. doi: 10.1016/j.bpj.2021.04.013